Artificial Substrates for Analysis of Bacterial Biofilms in Aquifers of Mammoth Cave and Characterization of Several Bacterial Communities
It was shown in a previous post that the clastic sandy sediments found throughout the base-level streams of the Mammoth-Flint Ridge Cave System contained the highest concentrations of bacterial DNA, and these sediments were widely distributed throughout the world's longest cave system at Mammoth Cave National Park (MACA). The 16S rDNA of the sandy cave sediment eubacterial community at one important site called Charon's Cascade was quantified and analyzed by quantitative Real Time PCR (qRT-PCR), molecular cloning, DNA sequencing, and phylogenetic analysis in another previous post.
In order to study bacterial communities throughout the cave system and to study other cave systems, an artificial substrate was needed to eliminate the influence of geological variables such as rock type, particle size, and surface area. Our initial survey of potential study sites throughout the Mammoth Cave system showed a diversity of different sediments ranging from fine silt to small pebbles.

Photos by Rick Olson, Gary Berdeaux, and John Andersland.
However, some sites of interest were not composed of this ideal natural substrate, and one would not expect to find the same substrate in cave systems throughout the USA National Park Service (who funded this research) or elsewhere in the world. In order to normalize quantitative data to be expressed as units per gram of substrate, a universally equivalent artificial substrate was required.
BioSep® Beads
An artificial and homogeneous substrate was developed along with Dr. Kerry Sublette (University of Tulsa) and the late Dr. David C. White (University of Tennessee).
Small beads composed of 30% St. Louis limestone, quarried just outside park boundaries, were formed by fusion of Nomex® with approximately 30% micronized limestone (or CaCO3 labeled with the stable isotope carbon-13) at high temperatures with Nomex® to produce small BioSep® beads approximately 3 mm in diameter.
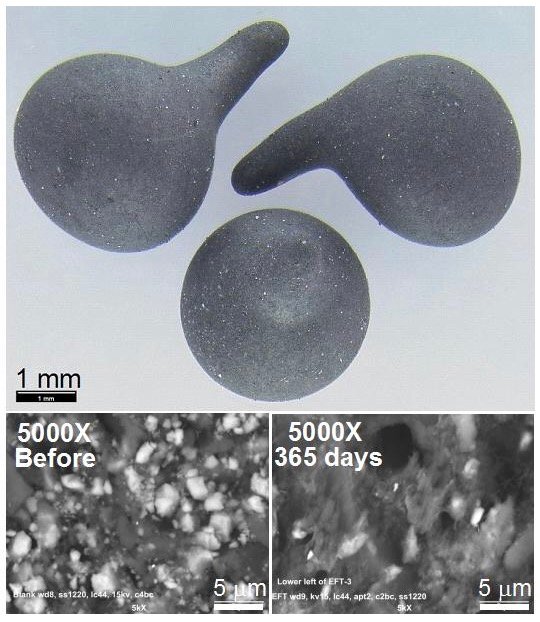
Photos by John Andersland, WKU.
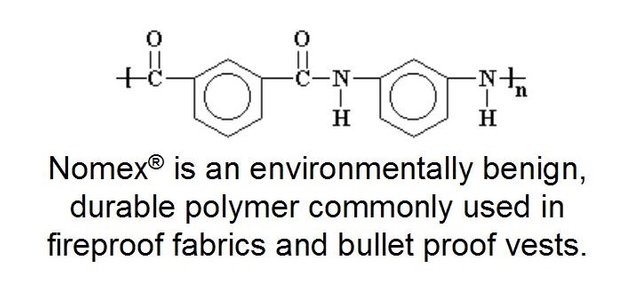
Diagram by @qiyi.
At each study site, six mesh dye trace bags containing approximately 3 grams (100 count) of the beads were secured with high tensile strength fishing line. Triplicate bags at each site contained BioSep® beads composed of either limestone or carbon-13 labeled CaCO3. The bags remained in place for one year during which time a bacterial biofilm developed on the bead surfaces (visible in the electron micrographs after one year in the cave). Samples were removed from the cave on ice and taken to the Biotechnology Center at Western Kentucky University (WKU) for analysis.
Clearly a thick covering of biofilm growth covers the limestone particles and the entire bead surface as seen in the Scanning Electron Micrographs (SEMs) provided by John Andersland, Dept of Biology, WKU. Environmental bacteria rarely live alone on a petri plate because they are part of an interdependent and delicate ecosystem. Small invertebrates survive in the absence of any apparent food source by grazing on the microbial biofilms invisible to the naked eye. They form the base of the entire cave food pyramid.

Photo by @qiyi and Rick Olson.
Hypothesis and Experimental Approach
We already knew that Terminal Restriction Fragment Length Polymorphism (TRFLP) profiles clearly identified a particular eubacterial 16S rDNA similar to Nitrospira moscoviensis. It was found throughout the Mammoth Cave karst aquifer and also in other caves in Kentucky, Tennessee, Alabama, Arkansas, and Missouri.
If it and similar kinds of nitrite-oxidizing bacteria were contributing to cavern enlargement and limestone dissolution by utilizing inorganic carbon from limestone (CaCO3) in the extremely nutrient limited conditions of the cave, then it would be expected that carbon-13 would be incorporated into the biomolecular makeup of the bacteria growing on the isotopically-labeled beads. The technique to demonstrate this was to be Phospholipid Fatty Ester Analysis (PLFA) that relies on mass spectrometry to identify and characterize membrane lipids.
Half of each bag of limestone beads was reserved for characterization of bacterial communities by quantitative PCR and phylogenetic analysis of 16S rDNA sequences, while the other half was sent to the University of Tennessee for PLFA analysis. All carbon-13 labeled beads were sent for PLFA analysis.
PLFA results for the limestone beads will be described in a later post, and the DNA results will be discussed here. Unfortunately, Dr. White passed away during the study and the carbon-13 labeled beads were never analyzed. The PLFA values reported below are an accurate measure of total biomass, both prokaryotic and eukaryotic.
Introducton and Significance
The caves and underground aquifers of the South Central KY karst region and the Mammoth-Flint Ridge system, with over 640 km (400 miles) of surveyed passages and by far the longest known cave on Earth, are the focus of many interdisciplinary studies. Mammoth Cave National Park (MCNP) is a Mecca for archaeologists, biologists, cavers, geologists, hydrologists, speleologists, and tourists from all over the world. Dual designations as an International Biosphere Reserve and a World Heritage Site testify to its unique biological diversity and cultural significance.
The Mammoth-Flint Ridge cave system and its associated watershed is the premier example of Earth’s karst aquifers, in which dissolution of soluble limestone bedrock plays a key role in the development of porosity and permeability. Globally, karst aquifers underlie some 12% of the terrestrial land surface and have been estimated to supply drinking water to 25% of the world's population. In the USA, 20% of the land surface is karst and 40% of the groundwater used for drinking comes from karst aquifers.
Limestone caves represent groundwater conduits that either have been abandoned by flowing water or are actively enlarging due to dissolution by flowing water. Indeed, Mammoth Cave provides an immense underground laboratory in which to study groundwater aquifer dynamics of importance for global society, and to assess the impact of human civilization on aquifer evolution and water quality.
Study Design and Supervision
In order to implement this study for the National Park Service (NPS), all NPS activities and scientific contributions were made possible by and belong to Rick Olson, Chief Ecologist at MACA. Ranger Olson selected all study sites, coordinated and guided all cave excursions, and handled all executive matters regarding contracts, permits, and funding. Ranger Olson, along with Horton Hobbs, has published a book that includes some of the data presented here. "Mammoth Cave: A Human and Natural History" recently hit the bookshelves.
This Unpublished Data was handed over to the MACA Division of Science and Resource Management (SRM) by 2007 for the Long Term Monitoring Program (LTMP), and NPS funding for the project from 2001 to 2006 was implemented through the National Cave and Karst Research Institute (NCKRI)
Eventually five sites were selected for the study by Ranger Olson. His originally authored complete list of potential monitoring sites can be seen here. DNA analysis was carried out by @qiyi, with the help of many graduate and undergraduate student assistants, essentially as described in a previous post. With artificial substrates, one gram (about 30 each) of BioSep® beads was used with a 10 g soil DNA extraction kit (MOBio Laboratories, Carlsbad, CA) instead of one gram of sediment with a one gram kit. The final product was isolated in 1 milliliter instead of 50 microliters. A negative control consisted of DNA extracted from beads that had never left the laboratory.
All experiments with qRT-PCR were carried out as described previously using DNA extracted from beads instead of DNA extracted from sediments. Cloning, sequencing, and phylogenetic analysis were carried out as described in a previous post.
Results
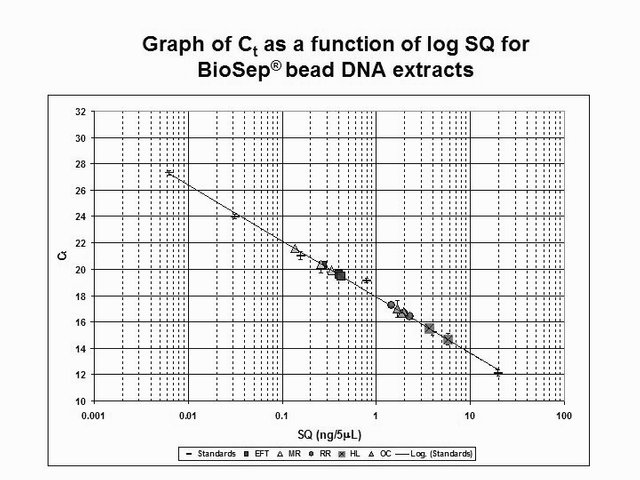
Diagram by @qiyi.
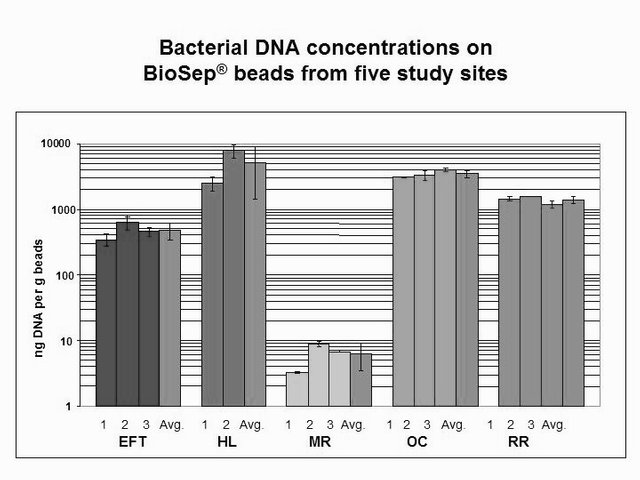
Diagram by @qiyi.
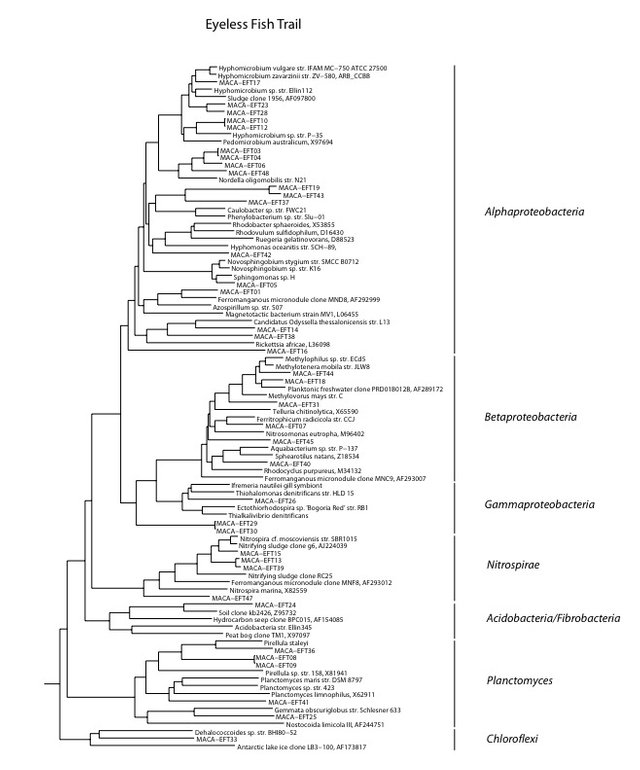
Eyeless Fish Trail (EFT)
This site is one of the most protected habitats for the Kentucky Cave Shrimp. A healthy population of shrimp can usually be seen at the site, and it can only be reached in low water conditions.
We found a lower than average biomass of bacterial DNA with six major divisions represented at the phylum and subphylum level among 38 total clones. The site is relatively unimpacted and presumably the healthy shrimp population is an indication that this is an ideal example of a cave aquifer. The data are tabulated below, and summarized in the accompanying diagrams:
N = 38 clones
476 ng bacterial DNA per gram of beads (+/-17%)
726 pmol PLFA per gram of beads (total biomass)
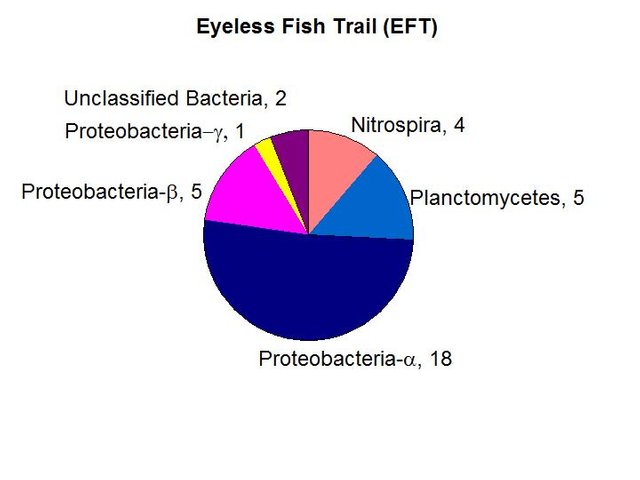
Results from taxonomic classifications using the RDP. Diagram by @qiyi
Phylogenetic tree by Hazel Barton.
Link to PDF
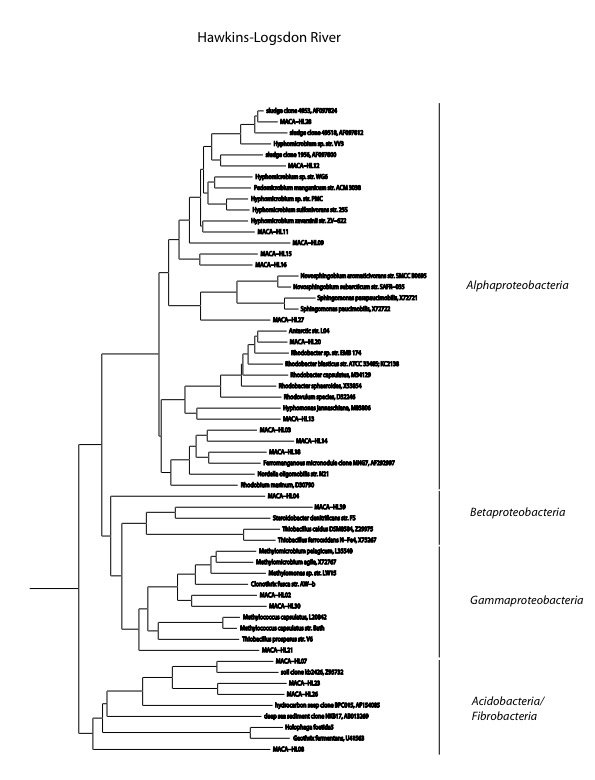
Hawkins-Logsdon Confluence
This site is like the iliac arterial branch of the cave system. The two largest inputs from Cave City to the north and Park City to the south converge at this point.
The combined flow constitutes the main aquifer of the Mammoth Cave System. It can only be visited during low flow, and presumably the bead bags left there experienced a tremendous amount of turbulence from time to time. One of the three bags was lost, so we have a duplicate instead of a triplicate for analysis.
Amos Hawkins formation and Hawkins-Logsdon River site, Photos by Gary Berdeaux (2006).
We found a the highest biomass of bacterial DNA with six major divisions represented at the phylum and subphylum level among 22 total clones. The site receives major drainage from the north and the south to converge and flow through the Historic Section of Mammoth Cave itself. The data are tabulated below, and summarized in the accompanying diagrams:
N = 22 clones
5135 ng bacterial DNA per gram of beads (+/-51%)
1370 pmol PLFA per gram of beads (total biomass)
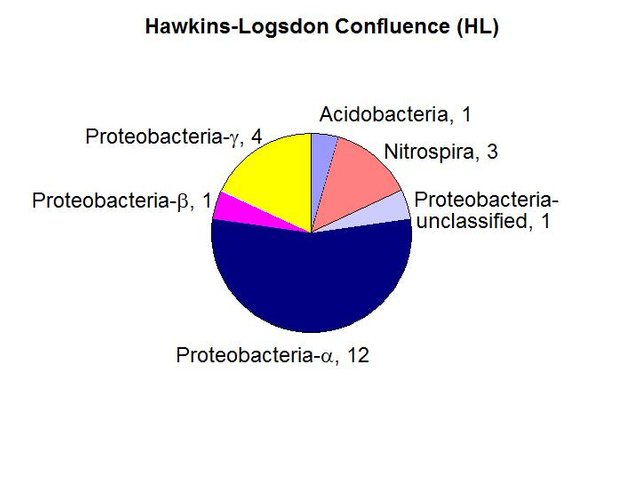
Results from taxonomic classifications using the RDP. Diagram by @qiyi.
Phylogenetic tree by Hazel Barton.
Link to PDF
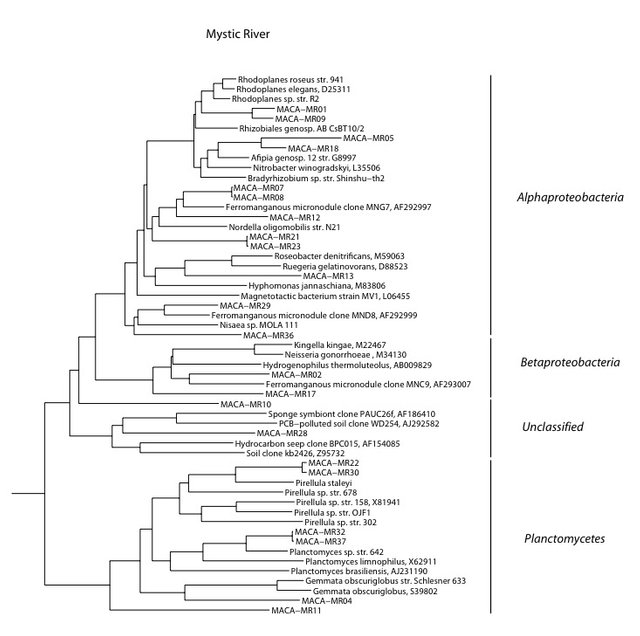
Mystic River (MR)
River Midstream is close to being a pristine cave stream. There is a substantial population (relatively speaking) of the endangered Kentucky Cave Shrimp in this area, and populations of other aquatic cave fauna appear to be normal in numbers and age distribution. The shrimp are grazers, and presumably are consuming the type of microbial biofilm they developed with and are adapted to. Olson’s hypothesis was that extreme energy and nutrient limitations in a minimally impacted cave stream are the determining factors shaping an oligotrophic microbial community.
We found the lowest biomass of bacterial DNA with six major divisions represented at the phylum and subphylum level among 26 total clones. The site has the most hypotrophic and pristine setting of all those studied. It receives drainage from essentially virgin forest on the surface. The data are tabulated below, and summarized in the accompanying diagrams:
N = 26 clones
6.3 ng bacterial DNA per gram of beads (+/-26%)
956 pmol PLFA per gram of beads (total biomass)
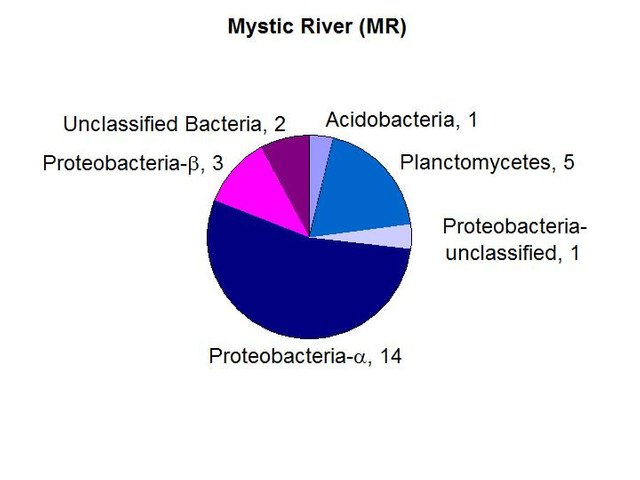
Results from taxonomic classifications using the RDP. Diagram by @qiyi.
Phylogenetic tree by Hazel Barton.
Link to PDF
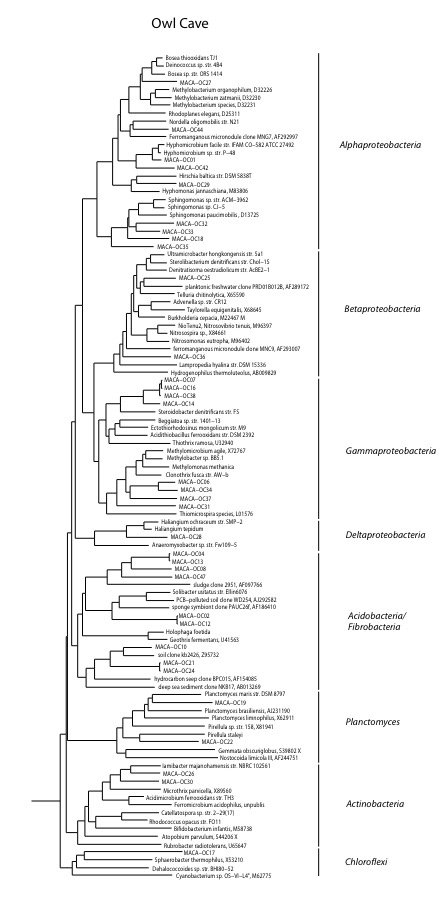
Owl Cave (OC)
Owl Cave is far downstream in the Turnhole Spring Basin, which extends far beyond park boundaries in the Sinkhole Plain with agriculture, oil and gas wells, Interstate Highway 65, and the CSX Railroad. Therefore contaminants of several types pass through the stream in Owl Cave. Natural backflooding events from Green Rver, and events caused by dam releases upstream are also a factor. For many years Olson has observed a heavy biofilm on rocks in the stream, which suggests organic enrichment. There is also a lack of cave adapted life, which might be partially accounted for by daylight hitting the stream, but even surface isopods have been absent so toxins may be at least sporadically present.
Owl Cave, Mammoth Cave Nat'l. Park, Photo by Chris Groves.
We found a relatively high biomass of bacterial DNA with eight major divisions represented at the phylum and subphylum level among 34 total clones. The site is clearly contaminated and is just hundreds of feet below a poultry farm near the southern park boundary. The PLFA biomass is the highest of all sites examined. The data suggested the presence of a higher number of eukaryotes and the exposure to sunlight does allow for some photosynthesis. The data are tabulated below, and summarized in the accompanying diagrams:
N = 34 clones
3463 ng bacterial DNA per gram of beads (+/-7%)
2849 pmol PLFA per gram of beads (total biomass)
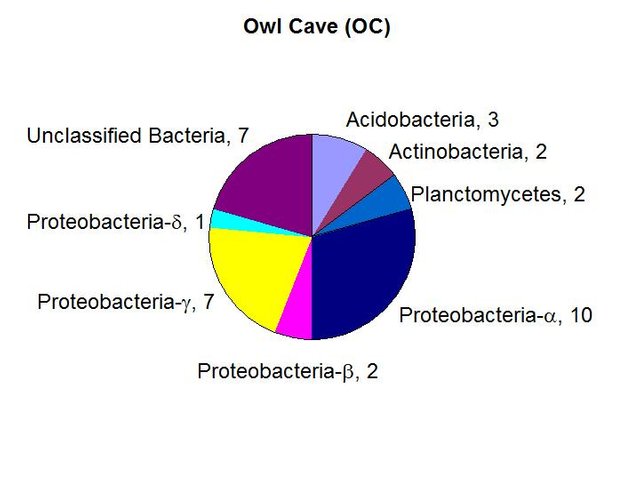
Results from taxonomic classifications using the RDP. Diagram by @qiyi.
Phylogenetic tree by Hazel Barton.
Link to PDF
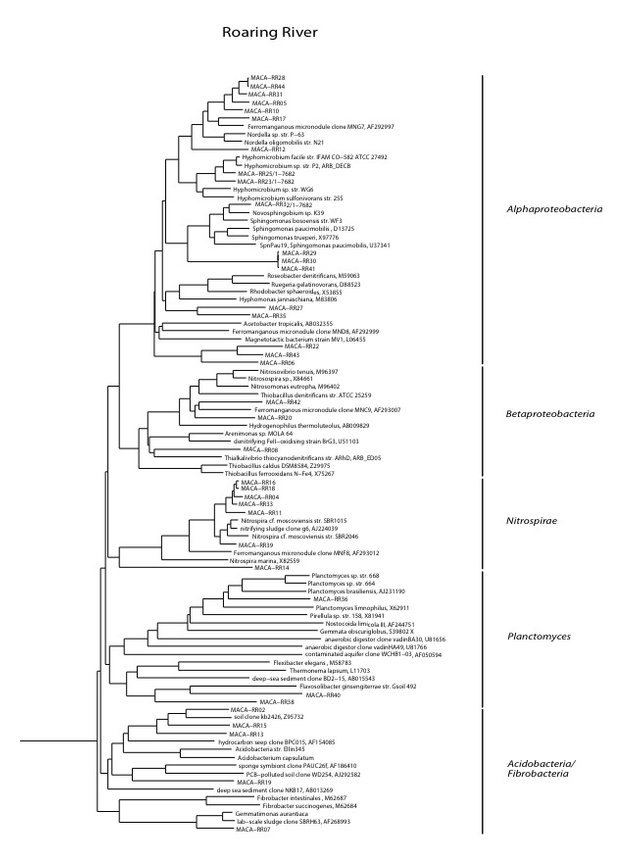
Roaring River (RR)
Roaring River Downstream is part of critical habitat for the endangered Kentucky Cave Shrimp designated by the U.S. Fish and Wildlife Service. This cave stream is thought to be moderately impacted because of flood overflow from the Turnhole Spring Basin. Olson’s hypothesis was that intermittent eutrophic conditions here are due to episodic organic enrichment caused by overflow events, which result in a microbial community intermittently eutrophic but generally oligotrophic, depending upon the frequency of overflow events.

Photo by Rick Olson.
We found a medium biomass of bacterial DNA with nine major divisions represented at the phylum and subphylum level among 36 total clones. The site is highly diverse and may be contaminated with fecal anaerobes. The data are tabulated below, and summarized in the accompanying diagrams:
N = 36 clones
1398 ng bacterial DNA per gram of beads (+/-8%)
441 pmol PLFA per gram of beads (total biomass)
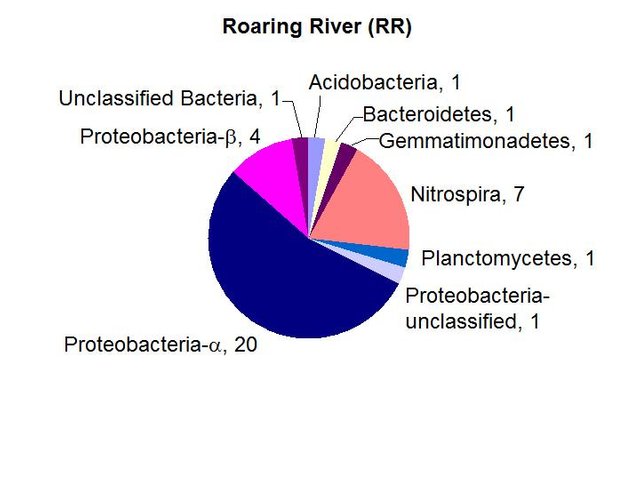
Results from taxonomic classifications using the RDP. Diagram by @qiyi.
Phylogenetic tree by Hazel Barton.
Link to PDF
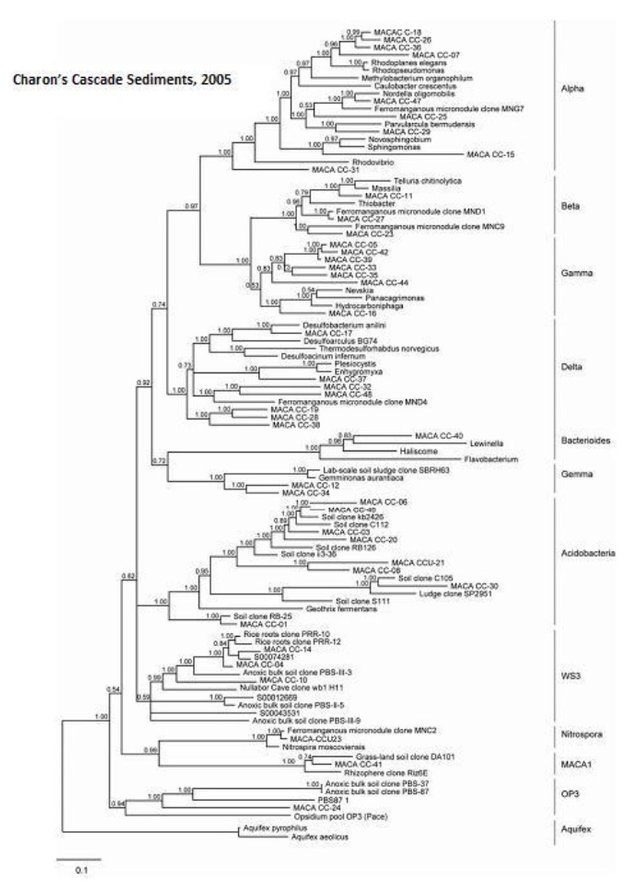
Charon's Cascade Sediments
For comparison here are the results from a previous post about bacterial communities in sandy sediments at Charon's Cascade.
We found a medium biomass of bacterial DNA with 10 major divisions represented at the phylum and subphylum level among 45 total clones. The site is highly diverse and may be contaminated with fecal anaerobes. The data are tabulated below, and summarized in the accompanying diagrams:
N = 45 clones
1261 ng bacterial DNA per gram of sediment (+/- about 10%)
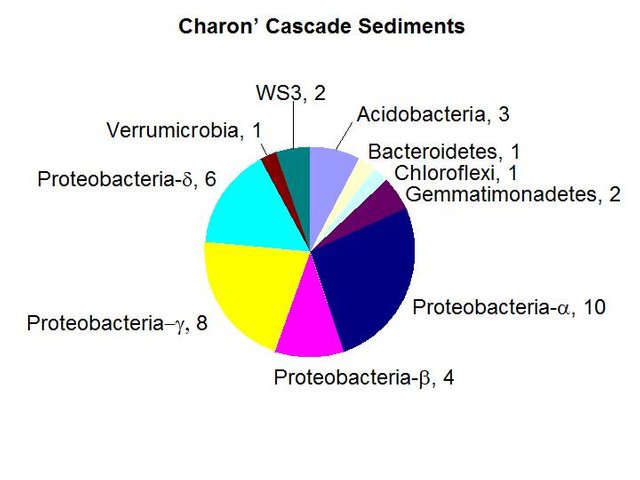
Results from taxonomic classifications using the RDP. Diagram by @qiyi.
Phylogenetic tree by Hazel Barton.
Link to jpeg photo of PDF
Conclusions
We conclude that the cave bacterial communities without toxic or outside influences maintains a steady rate of limestone dissolution not accounted for by geochemical and physical parameters. The microbial contribution is attributable to chemoautotrophic nitrite oxidizing bacteria where organic carbon is extremely limited. Microbial metabolism converts mineral carbonates into organic carbon, CO2 and aqueous bicarbonate, which can be converted to bacterial biomass while accelerating limestone dissolution.
Interdependence between nitrifying and denitrifying bacteria are fundamental to regional biomineral cycling of nitrogen and carbon in subterranean karst aquifers and caves in the Mammoth-Flint Ridge System. Knowledge of the bacterial community structure and estimates of bacterial biomass will provide baseline data needed for monitoring sensitive cave ecosystems and assessing their capacity for remediation.
Many of the 16S rDNA sequences suggest a role in bacterial iron-manganese redox as a possible energy source. Deposits of iron and manganese oxides are a prominent feature along virtually any route through the cave system.
The status of the bacterial communities at these five sites and at Charon’s Cascade in the year 2005-2006 is preserved in this molecular analysis. Incidents inside and outside park boundaries, along Interstate-65, agricultural activities on the sinkhole plain outside park boundaries to the south where population, transportation, and industrial growth is ongoing all will inevitably have an impact on the health of the cave system.
This study came to an end before November 3, 2007 when a construction project on the surface caused the release of E. coli into Charon’s Cascade, just beneath the visitors’ center. The National Park Service press release can be read here. Since then, a new drainage system and a new vistors’center have been built and the cave is a recommended place to visit for anyone!
Previous Posts in this Series
1. Introduction
2. Quantitative PCR: a brief introduction
3. Initial survey of various types of cave sediments
4. Quantification and characterization of bacterial DNA in Charon's Cascade Sediments
BioSep Bead References
Kerry L. Sublette, Sarkeys Professor of Environmental Engineering, University of Tulsa, 600 S. College Ave., Tulsa, OK 74104, (918)631-3085 Phone, (918)631-3268 Fax, [email protected]
Kerry L. Sublette, William A. Redman, Thomas I. Bair (2002). Adsorbent biocatalyst porous beads, US Patent # 6471864, The University of Tulsa, OK.
David C. White, Julia S. Gouffon, Aaron D. Peacock, Roland Geyer, Anita Biernacki, Greg A. Davis, Marsha Pryor, Mary Beth Tabacco, Kerry L. Sublette (2003). Forensic Analysis by Comprehensive Rapid Detection of Pathogens and Contamination Concentrated in Biofilms in Drinking Water Systems for Water Resource Protection and Management. Environmental Forensics 4 (1), 63-74.
M. Kästner, A. Fischer, I. Nijenhuis, R. Geyer, N. Stelzer, P. Bombach, C. C. Tebbe, H. H. Richnow (2006). Assessment of Microbial In Situ Activity in Contaminated Aquifers. Engineering in Life Sciences 6 (3), 234-251.
Kerry Sublette, Aaron Peacock, David White, Greg Davis, Dora Ogles, David Cook, Ravi Kolhatkar, Dennis Beckmann, Xiaomin Yang (2006). Monitoring Subsurface Microbial Ecology in a Sulfate-Amended, Gasoline-Contaminated Aquifer. Ground Water Monitoring & Remediation 26 (2), 70–78.
Other References
Angers, E.R., Northup, D.E., Reyesenbach, A.-L., Peek, A.S., Goebel, B.M., & Pace, N.R., 1998, Molecular phylogenetic analysis of a bacterial community in Sulphur River, Parker Cave, KY: American Mineralogist, v. 83, p. 1583-1592.
Barnes, S.M. & Nierzwicki-Bauer, S.A. 1997, Microbial diversity in ocean, surface, and subsurface environments: in J.F. Banfield and K.H. Nealson, (eds.), Geomicrobiology: Interactions between microbes and minerals, Reviews in Mineralogy: American Minerological Society, Washington, DC, v. 35 p. 35-71.
Barton, H.A., Taylor, M.R., & Pace N.R., 2003 phylogenetic analysis of a bacterial community in an oligotrophic cave environment: Geomicrobiology Journal, v. 21, p. 11-20.
Brucker R.W., Hess, J.W., &. White, W.B., 1972, Role of vertical shafts in the movement of ground water in carbonate aquifers: National. Speleological Society Bulletin, v. 10, no. 6, 9 pp.
Burrell P.C., Keller J., & Blackall L.L., 1998, Microbiology of a nitrite-oxidizing bioreactor: Applied and Environmental Microbiology, v. 64, no. 5, p. 1878-1883.
Dionisi, H.M., Layton A.C., Harms, G., Gregory, I.R., Robinson, K.G., and Sayler, G.S., 2002, Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira species from full-scale wastewater treatment plants by competitive PCR, Applied and Environmental Microbiology, v. 68, no. 1, p. 245-253.
Dowling, N.J.E., Mittelman, M.W., & White, D.C., 1991, The role of consortia in microbially influenced corrosion: in J. G. Zeikus, (ed.), Mixed Culture in Biotechnology, McGraw Hill, NewYork, NY, p. 341-372.
Elliott, L., Wright, S., Coakley, T., & Groves, C., 2000, Microbial ecology of conduit stream interstital fluids of the southcentral Kentucky karst aquifer: Impacts on aquifer development: Proceedings of Mammoth Cave National Park's Eighth Science Conference, p. 57-60.
Ehrich, S., Behrens, D., Lebedeva, E., Ludwig, W., & Bock, E., 1995, A new obliagtely chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscovienses sp. no. and its phylogenetic relationship: Arhives of Microbiology, v. 164, p. 16-23.
Fliermans, C.B., & Schmidt, E.L., 1977, Nitrobacter in Mammoth Cave: International Journal of Speleology, v. 9, p. 1-19.
Ford, D.C. & Ewers, R.O., 1978, The development of limestone cave systems in the dimensions of length and depth: Canadian Journal of Earth Science, v. 15, p. 1783-1798.
Ford, D.C. & Williams, P., 1989, Karst geomorphology and hydrology, Unwin Hyman Ltd., Winchester, MA, 601 pp.
Fowler, R., Roberson, E., and Sahi, S. (2003). Quantitative Real-Time Assays of Bacterial DNA in Sediments of the Flint-Mammoth Cave System with evidence for Nitrospira spp. at sites Undergoing Limestone Dissolution and Karst Aquifer Evolution. Proceedings of the 16th Annual National Cave and Karst Management Symposium, Gainesville, FL, Oct. 13-17.
GenBank, 9 Jul 2003, NCBI BLAST: National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/genbank
Groves, C. 2002. Personal communications.
Groves, C. & Meiman, J., 2001, Inorganic carbon flux and aquifer evolution in the South Central KY karst: U.S. Geological Survey Karst Interest Group Proceedings, St. Petersburg, FL, Feb. 13-26, 2001, Water-Resources Investigations Report, 01-4011, p. 99-105.
Groves, C. & Meiman, J., 1996, What are we learning at the Hawkins River Site? Proceedings of the Fifth Mammoth Cave Science Conference, p. 131-136.
Groves, C., Meiman, J. & Howard, A.D. 1999, Bridging the gap between real and mathematically simulated karst aquifers: Invited paper for the Proceedings of the Karst Waters Institute International Symposium on Karst Modeling, Charlottesville, VA, p. 197-202.
Harms, G., Layton, A.C., Gregory, I.R., Garrett, V.M., Hawkins, S.A., Robinson, K.G., & Sayler, G.S., 2003, Real-Time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant: Environmental Science and Technology, v. 37, p. 343-351.
Hess, W.H. 1900, The origin of nitrates in cavern earths: Journal of Geology, v. 8, p 129-134.
Hill, C.A., 1981, Origin of cave saltpeter: National Speleological Society Bulletin, v. 43, p. 110-126.
Holmes, A.J., Tujula, N.A., Holley, M., Contos, A., James, J.M., Rogers, P., & Gillings, M., 2001, Phylogenetic structure of unusual aquatic formations in Nullarbor caves, Australia: Environmental Microbiology, v. 3, no. 4, p. 256-264.
Hugenholtz, P., Pitulle, C., Hershberger, K.L., & Pace, N.R., 1998, Novel division level diversity in a Yellowstone hot spring: Journal of Bacteriology, v. 180, no. 2, p. 366-376.
Lane, D.J., 1991, 16S/23S rRNA Sequencing: in E. Stackenbrandt and M. Goodfellow (eds), Nucleic acid techniques in bacterial systematics, John Wiley and Sons, Inc. New York, NY, p. 115-148.
Northup, D.E., Barns, S.M., Yu, L.E., Spilde M.N., Schelble, R.T., Dano K.E., Crossey L.J., Connolly, C.A., Boston P.J., Natvig, D.O., & Dahm, C.N., 2003, Diverse microbial communities inhabiting ferromanganese deposits in Lechuguilla and Spider Caves, Environmental Microbiology v. 5 no. 11 p. 1071-1086.
Northup, D.E., Dahm, C.N., Melim, L.A., Spilde, M.N., Cossey, L.J., Lavoie, K.H., Mallory, L.M., Boston, P.J., Cunninhamm, K.I., & Barns, S.M.,2000, Evidence for geomicrobial interactions in Guadalupe Caves: Journal of Cave and Karst Studies, v. 62, no. 2, p. 80-90.
Pace, N.R., 1997, A molecular view of microbial diversity in the biosphere: Science, v. 276, p. 734-740.
Palmer, A.N., 1981, A geological guide to Mammoth Cave National Park: Teaneck, N.J., Zephyrus Press, 210 pp.
Ponchel. F., Toomes, C., Bransfield, K., Leong, F.T., Douglas, S.H., Field, S.L., Bell, S.M., Combaret, V., Puisieux, A., Mighell, A.J., Robinson, P.A., Ingelehearn, C.F., Isaacs, J.D., & Markham, A.F., 2003, Real-Time PCR based on SYBR-Green I Fluorscence: An alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications, and micro gene deletions, BMC Biotechnology, v. 3, no. 18.
Rusterholtz, K.J. & Mallory, L.M., 1994 Density, activity, and diversity of bacteria indigenous to a karstic aquifer: Microbial Ecology, v. 28, p. 79-99.
Sambrook, J., Fritsch, E.F., & Maniatis, T., 1989, Molecular cloning, a laboratory manual (2nd edition): Cold Spring Harbor Laboratory Press, v. 1, p. 1.53-1.86.
Schramm, A., De Beer, D., Gieseke, A., & Amann, R., 2000, Microenvironments and distribution of nitrifying bacteria in a membrane bound biofilm: Environmental Microbiology v. 2, no. 6, p. 680-686.
Siering, P.L., 1998, The double helix meets the crystal lattice: The powers and pitfalls of nucleic acid approaches to mineralogical investigations: American Mineralogist v. 83, no. 11-12, Part 2, p. 1593-1607.
Stein, L.Y., La Duc,M.T., Grundl,T.J. & Nealson,K.H., 2001, Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments, Environmental Microbiology, v. 3, no. 1, p. 10-18.
Thrailkill, J., 1968, Chemical and hydrologic factors in the excavation of limestone caves, Geological Society of America Bulletin. v. 79, p. 19-46.
Vaughan, K., 1998, A quantitative analysis of interstitial fluid chemistry and limestone dissolution rates within the clastic sediment of a karst aquifer conduit, Mammoth Cave, KY, M.S. Thesis, Western KY University, 128 pp.
Vaughan, K., Groves, C., & Meiman, J. 1998, Carbonate chemistry and interstitial fluids within cave stream sediments: in Proceedings of the International Geological Correlation Program, Project 379: “Karst processes and the global carbon cycle,” Bowling Green, KY.
White, W.B., 1988, Geomorphology and hydrology of karst terrains: Oxford University Press, New York , NY, 464 pp.




Hello qiyi!
Congratulations! This post has been randomly Resteemed! For a chance to get more of your content resteemed join the Steem Engine Team
Congratulations @qiyi! You have completed some achievement on Steemit and have been rewarded with new badge(s) :
Click on the badge to view your Board of Honor.
If you no longer want to receive notifications, reply to this comment with the word
STOPCongratulations! this post got an upvote by @steemrepo and was manually picked by the curator @yanosh01 to be added on STEEM REPOSITORY, simply comment "YES" and we upload it on STEEM REPO Website.
Want to know more about the Steem Repo project? Contact us on Discord
"YES"
Price is what you pay. Value is what you get.
A very impressive article. Great work!
The original written report went missing and the accompanying web page was deleted for that whole project. I had to reconstruct almost everything.
I looked again at an earlier study by Fliermans & Schmidt (1977) that demonstrates the presence of chemolithotrophic nitrifying bacteria (Nitrobacter sp.) using species-specific fluorescent antibodies to show Nitrobacter cells attached to sediment particles associated with saltpeter mining:
http://scholarcommons.usf.edu/cgi/viewcontent.cgi?article=1493&context=ijs
Do you kno da wei?