Finding, Counting, and Ordering Genes Using Incredibly Sophisticated Biomolecular Megatechnology

The future is so close, it's already happened.
Source: Pixabay
Introduction
Science and technology are like two equally addicted alcoholics: they enable each other. In this post I will go over some scientific discoveries that would have happened much slower, if at all, without the help of some intricate and very high-tech and mind-blowing inventions.
So let's stop introducin' and let's get inventin'!
Sir Archibald Garrod
Source: in Wikimedia Commons contributors
We all know (well, biologists and biology-aficionados do!) about the Central Dogma: about how "DNA encodes RNA encodes proteins". But it wasn't always so. Mendel, who didn't even use the word gene, and talked instead about heritable factors, didn't venture a half-guess as to how genes are able to determine phenotypes. Sir Archibald Garrod, however, did.

For a medical doc, Garrod's signature is surprisingly legible.
Source: Wikimedia Commons contributors
Archibald Garrod was an English physician who worked at a children's hospital. With Mendelian inheritance in his toolbelt, it didn't take long before he realized certain diseases run in families. One disease that got his special attention was alkaptonuria, whose characteristic visible symptom is black urine, because of the presence in the urine of a molecule called alkapton. Garrod studied family trees, and realized the disease was autosomal recessive[2]: the child needed to inherit two "broken" alleles to get the disease (alleles are the two versions of a gene, one from mom and one from dad). Garrod hypothesized that the disease was due to a mutation in a metabolic pathway that rendered the patient unable to metabolize alkapton, which was then excreted in the urine.[2]
And that was about as far as it went. Until two other fellas came along.
Finding genes: Beadle and Tatum hunt mutants

No, they were not a comedy duo, though their names would fit the role quite well.
Source: Beadle (right) & Tatum (left), in Wikimedia Commons contributors, modified
George Beadle and Edward Tatum were non-comical biologists, who preferred mold to children. Or at least that's what they preferred to study in their lab. Neurospora crassa, you see, has advantages that children don't have, like quick life cycles and no one arresting you when you expose them to mutagens.
(For information on how to radioactively mutagenize your children, or anyone for that matter, read @proteus-h's instructive post here.)
Why did Beadle and Tatum want to mutagenize their mold? The answer is much similar to the reason I gave my parents and my disgruntled older brother when they asked me why I broke his toys and tore furniture coverings and other things: "I want to know how they work" and "I want to know what's in them".
What can I say, I was a budding scientist.
Similarly, Beadle and Tatum wanted to make mutants in order to study what genes did. It's like turning a switch off to see which light or house-function it controls.
So how did they do it?
First, you pick an organism that can grow on minimal media: just a carbon source, salts, and water. (Children can't do this. Neurospora can.) This is ideal because an organism that can grow on minimal media has metabolic pathways that lead from that minimal media to the amino acids and vitamins that are essential for cells to function. If you can make a mutant that has lost the ability to grow on minimal media, you can study that metabolic pathway.
Second, you treat them with a mutagen, like x-rays.[6] This increases the chances that a mutation will occur.
Three, plate the possibly mutant Neurospora on rich media before Professor Charles Xavier has had a chance to recruit them. (Rich media are the opposite of minimal. They are also called culture media, because you use them to culture your organisms to visible proportions to the naked eye. Culturing organisms =/= taking them to museums.)
Four, transfer them to minimal media using pin-point accuracy technology.

Do not try this at home. This advanced equipment must only be handled by trained professionals and people who just had lunch. Rule of thumb: if you don't have a biochemistry PhD, do NOT attempt. (If you still insist on being a Daredevil, read this article and make sure you sterilize your toothpicks.)
Source: Wikimedia Commons contributors
Five, see which cultures fail to grow. Those are your mutants.
Six, using the incredibly sophisticated technology cited above, transfer the mutants to various plates that contain minimal media plus some vitamin or amino acid. So, if you add valine (an amino acid), and the mutant still can't grow, it means that's not where the problem's at. If you then add arginine and the mutant grows, it means the mutation is in a gene that controls the synthesis of arginine from minimal media. The mutant has lost its ability to make arginine, so adding it enables the Neurospora to grow.
Dominant or recessive?
Sir Archibald Garrod (the black piss guy) determined dominance by studying family trees. These are somewhat lacking when it comes to Neurospora. They're not big on keeping records. So how do you determine whether a mutation is dominant or recessive?
Simple: you mate the mutant with a wild type (normal) spore. Then plug-and-play Mendel's laws of inheritance: if the mutation goes away in the next generation (= can grow on minimal media), it means it's recessive. If it stays, it's dominant.
Counting genes: Complementation[8]

You complement me.
Source: Pixabay
You know how two people in a relationship often have strengths that complement each other's weaknesses? Like one's sociableness complements the other's shyness, a wife's politeness complements her husband's crude nature, or a wife who is a spendthrift complements her husband's ability to make money, so much so he's hardly able to keep up? Well, something much similar happens at the genetic level, with what's called complementation.
Complementation is what you do when you try to determine which of the mutations isolated in the previous two sections of this post are the same mutations. So let's say you've isolated 100 mutants. Does that mean you got 100 different malfunctioning genes? Maybe some mutations are the same mutations. How can you tell?
Here's where complementation comes in. Complementation happens between two alleles. Alleles are two versions of a gene, and when one malfunctions the other can "rescue" it.
To get back to the "couples" analogy: a husband and a wife both have a defect (mutation). Is it the same defect? Let's say one defect is spending all one's money on clothes, the other defect is being a gambler. Both these defects will show up as bankruptcy (phenotype), so we don't know whether the underlying defect (mutation/gene) is the same. If we "mate" the two mutants together, then if both these mutants have the "spending all one's money on clothes" allele, the phenotype will be bankruptcy. Same if both have the gambling allele. But what if one mutant has the "clothes" allele and the other has the "gambler" allele? Well, when the couple go to the store, the gambler acts as a rational influence and prevents the other from spending all one's money. And when the couple go to the casino, the clothes-loving person is the one that now pulls the reins on the gambler. So the resulting phenotype is wild type: no bankruptcy! So we know we got two different genes at play.
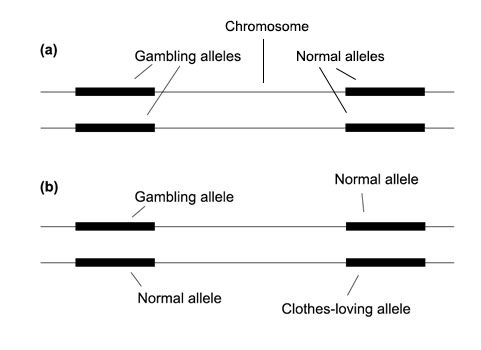
Here we see it pictorially. In case (a), the mating results in non-complementation — the result is still a mutant — which means we hit on the same mutation twice. In case (b), the mating results in a normal wild type, since each mutant complements for the deficiency of the other: the gambling gene is dominated by its normal counterpart allele, as is the clothes-loving allele.
Note that for this reason, this test can only be done with recessive phenotypes. That's why we had to distinguish between dominant and recessive in the previous step.
Here is a video that covers complementation tables, that I skipped here. Complementation tables are the Punnett squares of...complementation! They're cool, so I thought I'd mention them.
Ordering genes: Epistasis[9] [10]
You know there's some kind of pathway leading from the minimal media to the synthesis of a vitamin, or an amino acid. But how do you determine the order? You got 100 mutants, each one, say, representing a different gene. But what's the order of those genes? Does the gambling gene come before the clothes-loving gene, or after it?
Let's get more specific. Let's say we want to know how Neurospora makes Vitamin C. We know there's some kind of biochemical pathway leading from minimal media to Vitamin C production. Let's say, after much effort, we've only managed to find 4 mutants responsible for Vitamin C synthesis, and two of them were revealed by a test of complementation to be the same mutation, so we got a total of 3 Vitamin C mutants.
From this, we can hypothesize that there's a 3-step pathway leading from minimal media to Vitamin C. Let's represent this by letters A, B, and C. And we can use letters a, b, and c to represent the enzymes that will carry us along the pathway. These enzymes are coded by genes.
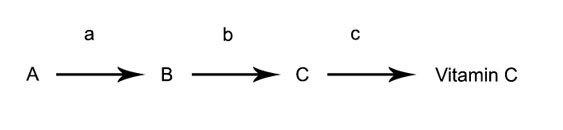
Now what if a mutant's mutation is in the gene that codes enzyme a? Would the mutant make Vitamin C if I gave it A? No, because it has a malfunctioning enzyme a that can't use A. But it would grow if I gave it any of the stuff in the later steps. The same isn't true for b, however. b wouldn't grow on A nor B. It would need C. And what about c? That would require straight-out Vitamin C in order to grow: it's the last step in the pathway, no supplementation can rescue it.
You might think we've cracked the puzzle, that we can now order genes. But this is actually quite laborious. There's often hundreds of steps in a pathway, and we often don't know if we got them all in our mutants. There's a more direct way to compare order.
Suppose I have two mutants, and I cross them with each other. Now I got a double mutant. Now what would that mutant look like? Say I cross mutant a with mutant b, what would it look like if I gave it the above test? It would look just like mutant b (= only C would rescue it). Mutant ab sort of "stands upon" the phenotype of mutant b. In other words, mutant ab is epistatic on mutant b. This is what's called an epistasis test. By crossing my two mutants, I know that the resulting phenotype will be the same as the phenotype of the mutant that is later in order.
Now I don't have to give hundreds or mutants hundreds of supplements in order to slowly figure out the order in which they come. I can do it two mutants at a time.
Don't get me wrong, the procedure is still laborious. Or at least it was, until a biologist came up with a new technology that made toothpicks a thing of the past.
Replica plating

Esther Lederberg, biotechnologist.
Source: Wikimedia Commons contributors
In a seminal paper, Esther Lederberg described a geometrically configurated, orientable, reliable, time-crunching, fiber-based, adaptively advantageous, densely inoculating, en masse technology, that would come to be known as replica plating.

I bet there's a curtain missing a piece somewhere in the building where Esther worked.
Source: Wikimedia Commons contributors
Basically it's an autoclaved (sterilized) velvet cloth secured on a block, and you use it by pressing a cultured plate on it, that may contain dozens of cultures, and then pressing on it new plates (that contain different media). You can literally transfer hundreds of colonies in a matter of seconds, whereas previously you'd have to do each single one by toothpick or other still-inadequate technology.
You can see replica plating being done live in this short video.
The scientific establishment was "appropriately in awe".[12] A fellow scientist described the innovative technology in these words:
It was brilliantly simple: creative discoveries often are. She thought of using ordinary velveteen from a yard goods store to serve as a kind of rubber stamp. The tiny fibers of the velveteen acted like hundreds of tiny inocculating needles. The pad was carefully kept in the same orientation and used to inocculate a series of agar plates containing different media containing antibiotics or supplemented with essential nutrients such as amino acids and vitamins. Esther and Joshua used this technique as an indirect selective method to prove the spontaneous origin of mutants with adaptive advantages.[12]
It's amazing what you can do with a piece of cloth and a block of wood.
Credits roll

It's rather astonishing, the things you can learn without needing any fancy machinery. Geneticists are in my opinion probably unsurpassed in this regard. They could find, count, and order genes without any DNA sequencing machinery, by employing the simplest of tricks, often simple logic tricks (like in the epistasis test case). In fact, they can do more than that, as has been seen in some of my previous posts, and as will be seen in my future ones.
In the tradition of a documentary's "where are they now?" trope after a movie ends, I want to briefly tell you what happened to our heroes above.
Much like Mendel, Garrod's work wasn't immediately recognized. And, again like Mendel, and in the same tradition of calling people "father of x" or "grandfather of y", Archibald became known as the "father of chemical genetics".[4]
Beadle and Tatum went on to win the 1958 Nobel Prize in Physiology or Medicine (to this day, no one knows which one), and in their acceptance speech they gave credit to Archibald Garrod, with a humility that is often a mark of true scientists:
[W]e had rediscovered what Garrod had seen so clearly so many years before. By now we knew of his work and were aware that we had added little if anything new in principle. [15]
Esther Lederberg went on to receive a number of professional honors, and was still occasionally seen in fabric stores searching for "the next big cloth" [reference pending].
References
1. Wikipedia contributors, "Central dogma of molecular biology," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Central_dogma_of_molecular_biology&oldid=815670496 (accessed January 4, 2018).
2. Wikipedia contributors, "Archibald Garrod," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Archibald_Garrod&oldid=813270862 (accessed January 4, 2018).
3. Wikipedia contributors, "Alkaptonuria," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Alkaptonuria&oldid=817774179 (accessed January 4, 2018).
4. 1908 Archibald E. Garrod (1857-1936) postulates that genetic defects cause many inherited diseases http://www.genomenewsnetwork.org/resources/timeline/1908_Garrod.php
5. Wikipedia contributors, "Neurospora crassa," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Neurospora_crassa&oldid=810148286 (accessed January 4, 2018).
6. G. W. Beadle and E. L. Tatum. 1941. Genetic control of biochemical reactions in Neurospora. PNAS, Vol. 27. http://www.pnas.org/content/27/11/499.full.pdf
7. Wikipedia contributors, "Growth medium," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Growth_medium&oldid=816061706 (accessed January 4, 2018).
8. Griffiths AJF, Miller JH, Suzuki DT, et al. An Introduction to Genetic Analysis. 7th edition. New York: W. H. Freeman; 2000. A diagnostic test for alleles. Available from: https://www.ncbi.nlm.nih.gov/books/NBK21921/
9. Griffiths AJF, Miller JH, Suzuki DT, et al. An Introduction to Genetic Analysis. 7th edition. New York: W. H. Freeman; 2000. Gene interaction and modified dihybrid ratios. Available from: https://www.ncbi.nlm.nih.gov/books/NBK21850/
10. Avery L, Wasserman S. Ordering gene function: the interpretation of epistasis in regulatory hierarchies. Trends in genetics : TIG. 1992;8(9):312-316. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3955268/
11. Huang, L. S. and Sternberg, P. W. Genetic dissection of developmental pathways (June 14, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.88.2, http://www.wormbook.org
12. Wikipedia contributors, "Esther Lederberg," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Esther_Lederberg&oldid=816796906 (accessed January 5, 2018).
13. Lederberg J, Lederberg EM. REPLICA PLATING AND INDIRECT SELECTION OF BACTERIAL MUTANTS. Journal of Bacteriology. 1952;63(3):399-406. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC169282/
14. Wikipedia contributors, "Replica plating," Wikipedia, The Free Encyclopedia, https://en.wikipedia.org/w/index.php?title=Replica_plating&oldid=812736310 (accessed January 5, 2018).
15. GEORGE W. BEADLE. Genes and chemical reactions in Neurospora. Nobel Lecture, December 11, 1958. https://www.nobelprize.org/nobel_prizes/medicine/laureates/1958/beadle-lecture.pdf
Earlier Introduction to Biology episodes:
7: Christmas Disease — Yes, it's real, 100% scientifically proven!
6: The Most Famous All-Nighter in the History of Genetics
5: Mendel's Lucky Number Seven — The law of genetics that almost wasn't
4: How Cells Use Logic To Do The Impossible
3 : Armchair Science — The Discovery of Proteins' Secondary Structure
2 : How Cell Membranes Form Spontaneously
1 : Eduard Buchner: The Man Who Killed Vitalism
steemSTEM is the go-to place for science on Steemit. Check it out at @steemstem or browse the #steemSTEM tag or chat live at steemit.chat
Excellent job highlighting the advancements of famous geneticists who didn't even need to know the structure of DNA to make their discoveries. I'd also like to add that replica plating is still in use today in many modern labs (though its thought of as an outdated technique at this point).
I'm still not convinced Neurospora crassa is a better test subject than children though.
Beautifully written, informative, entertaining, such a wonderful job @alexander.alexis.
You've truly come a long way in your writing of these science history blogs. I personally enjoy this content and I think a lot of others would too. Get the word out and promote your work!
Many thanks man!
I try to improve constantly. And I try to read others' posts, as many as I can, out of authentic interest, and comment, and even when I don't have something to say I still comment to let them know I've actually read the whole piece cos it makes writers feel good knowing that. It's only lack of time that's keeping me from doing more. But I'll get better!
But most kudos go to you! You're the engine behind the success of many steemians here! You don't know how many people's lives you're helping!
Well I for one have been enjoying these science history blogs too. Keep them coming man.
So true. So to honor that spirit I'm going to resteem this article. :-)
So true man... I agree with you. I was moved by that post @Alexis
I agree to this statement, DNA in the coming future can be altered there somehow a negative on it
As a microbiologist. This project is actually dangerous. It can lead the patient to be exposed to certain radiations. It looks promising. Just like @suesa said. "Promising isn't cure"
What are you referring to?
Hey, great post! We have featured your article in our weekly newsletter as #1 of last week! Keep up the good work, and let us know if you have any concerns.
Join us on the science-trail Discord
Cheers! )
Well done, sir! Up-voted and followed. Can't wait to read some of the others.
Are you familiar with http://genecoin.me/index.html ? Do you think this kind of tech is advanced enough to do what they say it can do?
That's interesting! But most of it sounds like bull, almost like they're trying to create digital versions of us. Sure you could store your DNA sequence in some kind of crypto, but who would pay for the sequencing?! And how exactly will they store the data on bitcoin?! It sounds like a scam overall, but I like the basic idea of people storing and sharing their DNA sequences, which will help medicine. You could give that data for free or sell it to bio companies, but you got datum for that.
I appreciate your professional insight. This topic is so way out of my leauge that its not even funny. I do think it might be useful to help prevent ID theft or be used to make some kind of personal ID that contained medical or financial information. Thanks again. Im glad you are on Steemit. I look forward to reading more of your stuff.
I'm hardly a professional myself!
Appreciate your stopping by!
It is known that Christmas disease is a commonly used term with the use of haemophilia B together.
I thought it was a shopping frenzy. But I learned that it was a blood disease.
an interesting name
You commented under the wrong post, but yeah!
sorry,
Nice one brother... We need more post like this to make steemit a place of learning as well. I really love it. Look forward to see your next post...
Very entertaining and informative article. To think that a toothpick is such an advance technological tool! I can no longer look at toothpick the same way again! Also I find your study of allele very interesting. So they are the culprit for my love of meat and sweets! Apparently I have inherited the allele of "can't live without meat" and the "I'll die of diabetes" allele. Anyway, thanks again and have a nice day.
:)
the color is good
Lol! This is so blatantly spammy it's almost worth a vote.
Almost.
He just needed to misspell a bit.
Now THAT would be worth a vote!
This flew over my head I think, but I'll take your word for it!
You didn't like my joke?
:(
Oh btw, check out the pic here, closely...
Maybe I read too much into it, I thought colr and goob might be biology jargon or acronyms I'm unaware of! But maybe you were just saying at least he spelled correctly, unlike most spammers!