Airborne microbes Part 2: How many bacteria do we breathe in each day?
Happy New Year Everyone!
In December, I began researching scientific literature to write a post about airborne microbes. I wanted to describe the sources of them, their potential health hazards, the quantity of microbes in the air we breathe, and how scientists can answer such questions. However, I realized that I delved into a topic that was too large for a single post. Therefore, I decided to split the post into two sister articles: The first was released a few weeks ago and describes the sources and health hazards of airborne microbes. The second (and final) in this short series is below.
In this post, I want to try to answer an intriguing question: How many bacteria do we inhale daily? As a microbiologist, I’m all too aware that we’re surrounded by airborne microbes. Whenever I work with in the lab, I have to rigorously apply what’s known as “sterile technique,” a set of standards and practices designed to minimize environmental contamination. Even when practicing sterile technique I find my media, reagents, and tools become contaminated all too often. Given this issue, I often wonder how many microbes are floating around the lab.
How do we assess airborne microbes?
Before we determine the number of bacteria we inhale, it is necessary to understand the different ways we can assess airborne microbe levels. Scientists have developed a number of ways to collect bacteria and fungi that are dispersed in the air, each with varying levels of accuracy. Below, I’ve listed some of the more common techniques
Agar plate exposure (1):
This is the easiest method to perform. An agar plate is exposed to air for a specific amount of time. The plate is then closed, and any microbes that landed on the plate are allowed to grow into colonies for several days. Scientists can count the number of colonies that grow after a certain period of exposure to estimate the number of bacteria in the air and can analyze colony morphology to identify them. This method is quick, cheap, and can be useful to a lab by showing the probability that media or other chemicals will become contaminated if exposed to air.Unfortunately, this method is also quite inaccurate. The number of bacteria that have a chance to land on the dish can vary widely depending on air currents and the location of the plate. Additionally, not every microbe that lands on the plate will develop into a colony. Some might not be compatible with the growth media or might grow too slowly to become a colony. Thus, scientists needed to develop methods with a higher degree of accuracy.
Impingers (1, 2):
Impingers are devices that push air through a liquid medium designed to capture bacteria. The liquid can then be analyzed for bacteria either by microscopic analysis, culturing, or more advanced methods. This is more accurate than agar plating because you can regulate the volume of air sampled, but it is not perfect because the liquid medium can damage microbes.
Impactors (1, 2):
Impactors are similar to impingers in that they push air at a medium designed to capture bacteria. However, with this instrument, the media is solid instead of liquid. The media can come in multiple forms, such as an agar gel to allow bacterial cultures to develop, or a water soluble membrane designed to pool bacteria for qPCR. As with the impingers, this device is not perfect because the collection method can damage organisms.filtration (3):
With filtration, air is pumped through a membrane, such as polycarbonate or cellulose acetate. The filter membrane allows air and other small molecules to pass through, but is small enough to trap microbes. This technique is generally the most accurate, but it tends to be the least cost effective and difficult to use. As a result, it is mostly used by researchers rather than for routine air quality tests. Different sized membranes can be used to trap different sized microbes as well.
Of these available methods, I want to find a study that uses the filtration technique because that is the most accurate available. I’m also consciously deciding to focus on just bacteria in the air rather than all microbes (bacterial, fungal, and viral) to simplify things.
So how many microbes are in the air?
Alright, now for the fun part where we get to do a deep dive into a paper. The paper in question, “Total Virus and Bacteria Concentrations in Indoor and Outdoor Air” was written by the Marr Lab at Virginia Tech in 2015 (4). This lab is dedicated to studying air quality and the first author, Aaron J. Prussin, has a number of papers focused on airborne microbes.
As the title suggests, the goal of this work is to assess airborne microbe levels across different locations. The authors traveled to several areas (classrooms, houses, outdoor areas, etc.) and used the filtration method described above to collect bacteria and viruses. They then stained the filter with a dye called SYBRGold that becomes fluorescent when it interacts with DNA or RNA. They looked at the filter with fluorescence microscopy to detect patches of nucleic acid. Each patch was assumed to indicate a bacteria or virus. Since a virus is much smaller than a bacterial cell, they were able to easily distinguish between the two types of organisms by size. Finally, the authors divided the number of organisms on the filter by the volume of air passed through the filter to calculate the airborne bacteria and virus concentrations in each location. The results are shown in the table below.
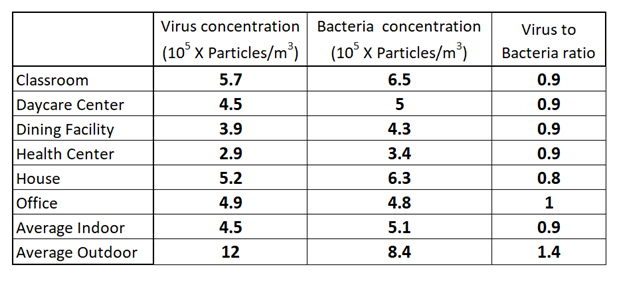
Airborne virus and bacteria concentrations in different environments. Keep in mind these numbers indicate tens of thousands per cubic meter. Adapted from (4).
In looking over this table, I noticed a few interesting observations. For one, indoor environments had similar levels of bacteria and virus particles. The most sterile appeared to be the health center, possibly because such areas are kept as clean as possible to prevent cross-contamination between sick individuals. I also noticed that the outdoors was significantly different from the average indoor space in that the total concentration of airborne microbes was higher and there was a greater proportion of virus particles compared to bacteria. Most importantly, this study is exactly what I needed because it gives us very clear numbers for the airborne bacterial concentrations indoors and outdoors. I believe that these numbers are accurate because they are close to the numbers determined by other similar studies (5, 6).
Before I move on, I wanted to mention a few other cool studies that analyze airborne microbes. Many studies are more concerned with the identity of airborne microbes rather than their quantities. In the paper “Chamber Bioaerosol Study: Outdoor Air and Human Occupants as Sources of Indoor Airborne Microbes” (7), the authors collected bacteria and fungi on a filter paper like in the previous study, but decided to identify them by DNA sequencing rather than quantitate them. Identification was accomplished by extracting DNA from the filter and sequencing all copies of the bacterial 16S rRNA gene and the ITS1 region of the fungal rRNA gene. These genes are highly conserved among related organisms and can be used to classify organisms into different taxa. I’ve described this technique in detail before so I’m just going to link to that description here.
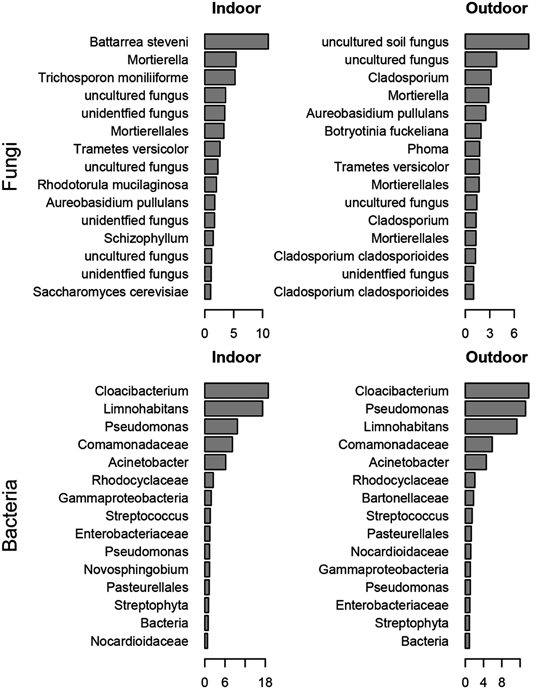
The relative abundance of the most common fungal and bacterial taxa in indoor and outdoor air Adapted from (7).
Ultimately, the authors were able to show that indoor and outdoor air contained different species of airborne microbes, and some taxa were abundant over others. For example, the fungi Battarrea steveni was abundant in indoor air samples, but barely detected in outdoor samples. However, they were unable to obtain quantitative data from this information.
One other study I want to mention is the “Stability of airborne microbes in the Louvre Museum over time” (8). There isn’t anything special about this paper from a scientific perspective. It uses typical methods to quantitate and identify bacteria in an enclosed space over the course of 6 months. It’s just the fact that this study is done in the most famous art museum in the world that fascinates me. Why would the authors want to know how many E. coli are floating around the Mona Lisa? Were petri dish cultures confused for fine art? Did the authors finally solve the Da Vinci Code? Well, I don’t have an answer to any of these questions because the full paper is locked behind a paywall.
So how many microbes do we breathe in each day?
Alright, so we have an approximate number for the concentration of bacteria in the air: 51,000 cells per cubic meter indoors and 84,000 cells per cubic meter outdoors. Again, I’m not including any virus or fungi in this count for the sake of simplicity. Now all we have to do is multiply this number by the volume of air we breath in a day to get the number of cells we inhale daily.
Determining the volume intake of an average human wasn’t too difficult. A quick google search revealed a study by the California Environmental Protection Agency (9) that focused on this exact question. I’m not sure why the Ca EPA needed to do this study, but it certainly helps me out. According to this study, an average adult male inhales about 8 liters of air per minute while resting. This comes to ~11,500 liters 11.52 cubic meters per day. Assuming this ideal man spends his entire day resting indoors, his bacterial intake will be…
51,000 bacteria/cubic meter X 11.52 cubic meters/day = 590,000 bacteria/day
590,000 bacteria per day!
Almost 600,000 bacteria per day, and that’s just if the guys sits down indoors all day playing video games. If he sat around outside his intake would increase to about 970,000 per day. That’s almost a million bacteria in a single day and the guy isn’t even walking around.
Now a million bacteria in a day seem like a huge number, but before you run out and buy a lifetime supply of facemasks, keep in mind a few things. Most airborne bacteria are harmless, not all inhaled bacteria would adhere to the lungs, and even fewer would be able to grow. Furthermore, one million bacteria pales in comparison to the estimated 100 trillion bacteria of the human microbiome (10). Still, I wouldn’t have guessed the amount of airborne bacteria we inhale each day to be as high as it is.
This project took a bit longer than expected, but I’m happy with the result. I’m also pretty confident in this number as multiple sources seem to back up the concentration of airborne bacterial and daily air intake of an average person. At any rate, I certainly learned an interesting fact and I hope you did too.
Images
All images used have been labelled for re-use by Wikipedia or are taken directly from the publication. If any image owner has an issue with this article, please contact me and I will address the issue.
Image Sources
IMG SRC 1: https://commons.wikimedia.org/wiki/File:Agar_Plate_Under_Sun
IMG SRC 2: https://en.wikipedia.org/wiki/Mona_Lisa
Sources
(1) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3444341/
(2) http://www.rapidmicrobiology.com/test-method/air-samplers/
(3) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3444341/
(4) https://www.ncbi.nlm.nih.gov/pubmed/26225354
(5) http://www.sciencedirect.com/science/article/pii/S0048969715308184#f0025
(6) https://www.sciencedirect.com/science/article/pii/S0021850217301891
(7) http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0128022
(8) https://www.ncbi.nlm.nih.gov/pubmed/23710880
(9) https://www.arb.ca.gov/research/resnotes/notes/94-11.htm
(10) https://en.wikipedia.org/wiki/Human_microbiota
About the Author
I’m a research scientist living in the suburbs of Boston. I recently left academia to work in in industry, but I miss teaching so I decided to start writing articles on interesting discoveries in the world of microbiology. My goal is to uncover subjects that are unusual, unique, and might one day have a major impact on our daily lives.
SteemSTEM
I’m a proud member of the @steemstem community and I encourage you to join. SteemSTEM is a community driven project which seeks to promote well written/informative Science Technology Engineering and Mathematics postings on Steemit. The project not only curates STEM posts on the platform through both voting and resteeming, but also re-distributes curation rewards as STEEM Power, to members of Steemit's growing scientific/tech community.
To learn more about the project please join us on steemit.chat (https://steemit.chat/channel/steemSTEM), we are always looking for people who want to help in our quest to increase the quality of STEM (and health) posts on our growing platform, and would love to hear from you!



What a blog about bacteria
I didn't know that we breath a million microbes every day that would be around 3 to 4 billion a year
That's insane
Yeah, way more than I expected. Thanks for reading!
You are welcome
To know about Cryptocurrency read my blogs
If you like them , let me know in the comments
I recently read some of this bacteria we absorb becomes part of our DNA, so you have now may DNA from someone who sneezed on you years ago!
Really that's awkward man really
Did you read any of my blogs?
Imagine how many more it'd be if viral material was included too :o
Yeah, adding viral particles and fungi would come close to tripling this number.
Yay? :/
Seriously pointless article about the Louvre. But... As there are people from all over the world, it would be cool if they made some fingerprint method for the estimation of the geographical origin of the visiors.
That would be pretty cool. If possible, it'd definitely be more interesting than the article they published.
That's really insane inhaling that much of micro bacteria!
Definitely. I would have guessed in the thousands, but not close to a million.
Ah, so I see the man in the first image sprays pollen when he sneezes! An inverse hay-fever of sorts.
Mixed pollen particles; SEM image
False-color SEM of chicory pollen
The numbers of bacteria we breathe daily are most likely over a million though ;)
Glad to see interest in microbiology!
Oops, thanks for catching the pollen issue. The title image is usually the last thing I put up and this time it appears I was focused on color scheme rather than scientific accuracy. I just replaced with a more accurate photo.
Regarding the number of bacteria we breath being likely over a million, is it just a hunch or do you see another error in my analysis?
Haha just poking a bit of fun. Looks like some Staphylococci on there now; fitting for a sneeze! Didn't expect the photo change, but cool -- attention to detail and annotation is a good thing to see in the SteemSTEM community :)
Re: bacterial numbers
Given that plate-based assays are often used as a metric for counting microbes, and we know that many microbes are viable but unculturable, it stands to reason that counting CFU on a plate will miss a lot of species that cannot grown on known mediums. Also, frequently bacteria exist on biofilms on a tiny mote of dust suspended in the air -- tiny to us, and so still likely to be counted as a individual CFU when in fact a colony frequently arises from many microbes. They are communal critters that play a numbers game, anyways.
As far as DNA sequencing goes, this assumes that all the 16s rRNA primers have the same priming efficiency and work on all bacteria -- including ones which have never had their genome sequenced, and may have a different enough rRNA region that fails to prime properly. I mean, we know what microbes we know and what tools work for them, but we honestly have no idea about most (>95%) microbes and only have fleeting glimpses of viable-but-not-culturable strains. It is also presumed that the polymerase used for PCR prior to sequencing is unaffected by methylation and DNA modifications, but that's getting more obscure.
There is also the confounding variable of our oral and naso-pharyngeal bacteria which are dividing and we inhale on a routine basis. There's also reference 1, which shows the millions of microbes that occupy the room with us, and multiply in our presence.
Many analyses are taken carefully with leaving collection methods relatively unperturbed; however, all the microbes and dust particles with attached biofilms or microbial aggregates that have settled on the floor will be stirred up by the draft of people's movement from room to room. This could be part of why outdoors -- where there is more biodiversity, but also breezes -- yields higher numbers of bacteria.
Good post though and breakdown of different methods/collection techniques! Glad to see more scientists on this platform :)
Ref:
1.) Qian, J., Hospodsky, D., Yamamoto, N., Nazaroff, W. W. and Peccia, J. (2012), Size-resolved emission rates of airborne bacteria and fungi in an occupied classroom. Indoor Air, 22: 339–351. doi:10.1111/j.1600-0668.2012.00769.x
Very well put and you even back it up with references. Consider me impressed:) I'd only point out that the study I referenced collected bacteria on a filter paper and counted individual cells via fluorescence microscopy. Its not a perfect method, but a little bit more accurate than counting CFUs. Aside from that, all your arguments are completely valid.
Its great to see another microbiologist on steemit. Consider me a follower from now on.
Great article, I always try to explain my patient about airborne transmission and this article is helpful
Glad you like it.
Cinnamon water with alcohol can be used to kill bacteria and pests using a spray.
Good advice.
Steemscience... SteemSTEM... This is very educational. I have learnt quite a few things about microbes through your post.... I guess billions x100 more microbes must have landed and proliferated on the mona lisa by now.
I support you with my resteem service
Your post has been resteemed to my 3300 followers
Resteem a post for free here
Power Resteem Service - The powerhouse for free resteems, paid resteems, random resteems
I am not a bot. Upvote this comment if you like this service
Hey abasinkanga. Thanks for the resteem! Glad you liked reading the post.
Yes. Its interesting... Knowing about the microbial lifeforms. Thanks for the upvote.
Do remember to use my resteem service again. Cheers!
Great reading of course! But learning that " Most airborne bacteria are harmless, not all inhaled bacteria would adhere to the lungs, and even fewer would be able to grow," I really would like to have more info on our relationships with this microbes.
Good point. I sort of made that assumption because the average human doesn't get sick every time they walk outside. Trying to demonstrate this from the available scientific literature would be a bit difficult and might constitute a post in itself, but I will look into it.
makes me want to purchase a hazmat suit for the hell of it, but honestly all of this is pretty accurate of course nothing in the universe can be trully mimicked or copied so some stats are just an estimate.
Yeah, actually knowing how many microbes are around and inside us can be very disconcerting. I'm hoping the estimate is accurate, but I could very well be under or over-estimating.