You are viewing a single comment's thread from:
RE: Airborne microbes Part 2: How many bacteria do we breathe in each day?
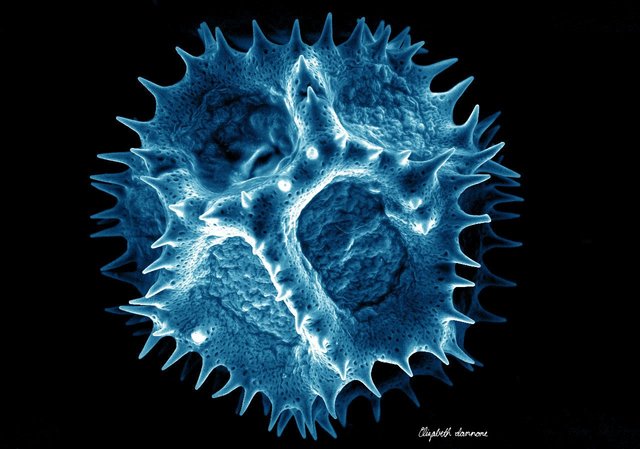
Ah, so I see the man in the first image sprays pollen when he sneezes! An inverse hay-fever of sorts.
Mixed pollen particles; SEM image
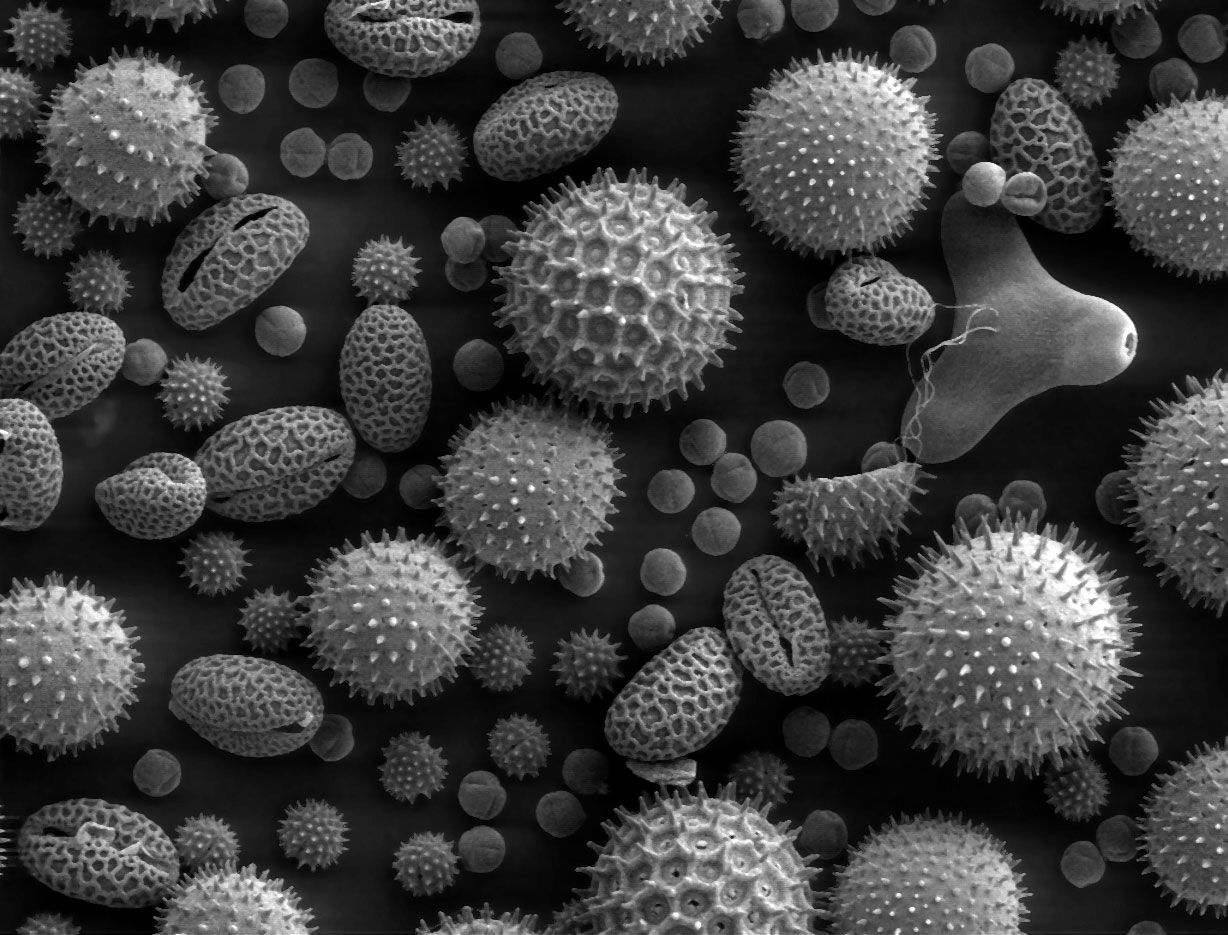
False-color SEM of chicory pollen
The numbers of bacteria we breathe daily are most likely over a million though ;)
Glad to see interest in microbiology!
Oops, thanks for catching the pollen issue. The title image is usually the last thing I put up and this time it appears I was focused on color scheme rather than scientific accuracy. I just replaced with a more accurate photo.
Regarding the number of bacteria we breath being likely over a million, is it just a hunch or do you see another error in my analysis?
Haha just poking a bit of fun. Looks like some Staphylococci on there now; fitting for a sneeze! Didn't expect the photo change, but cool -- attention to detail and annotation is a good thing to see in the SteemSTEM community :)
Re: bacterial numbers
Given that plate-based assays are often used as a metric for counting microbes, and we know that many microbes are viable but unculturable, it stands to reason that counting CFU on a plate will miss a lot of species that cannot grown on known mediums. Also, frequently bacteria exist on biofilms on a tiny mote of dust suspended in the air -- tiny to us, and so still likely to be counted as a individual CFU when in fact a colony frequently arises from many microbes. They are communal critters that play a numbers game, anyways.
As far as DNA sequencing goes, this assumes that all the 16s rRNA primers have the same priming efficiency and work on all bacteria -- including ones which have never had their genome sequenced, and may have a different enough rRNA region that fails to prime properly. I mean, we know what microbes we know and what tools work for them, but we honestly have no idea about most (>95%) microbes and only have fleeting glimpses of viable-but-not-culturable strains. It is also presumed that the polymerase used for PCR prior to sequencing is unaffected by methylation and DNA modifications, but that's getting more obscure.
There is also the confounding variable of our oral and naso-pharyngeal bacteria which are dividing and we inhale on a routine basis. There's also reference 1, which shows the millions of microbes that occupy the room with us, and multiply in our presence.
Many analyses are taken carefully with leaving collection methods relatively unperturbed; however, all the microbes and dust particles with attached biofilms or microbial aggregates that have settled on the floor will be stirred up by the draft of people's movement from room to room. This could be part of why outdoors -- where there is more biodiversity, but also breezes -- yields higher numbers of bacteria.
Good post though and breakdown of different methods/collection techniques! Glad to see more scientists on this platform :)
Ref:
1.) Qian, J., Hospodsky, D., Yamamoto, N., Nazaroff, W. W. and Peccia, J. (2012), Size-resolved emission rates of airborne bacteria and fungi in an occupied classroom. Indoor Air, 22: 339–351. doi:10.1111/j.1600-0668.2012.00769.x
Very well put and you even back it up with references. Consider me impressed:) I'd only point out that the study I referenced collected bacteria on a filter paper and counted individual cells via fluorescence microscopy. Its not a perfect method, but a little bit more accurate than counting CFUs. Aside from that, all your arguments are completely valid.
Its great to see another microbiologist on steemit. Consider me a follower from now on.