Predicted Labeling Of Calvin Cycle Intermediates With Carbon-13 Labeled Carbon Dioxide
In previous posts I have discussed the importance of isotopic labeling in metabolic pathway discovery, metabolic flux analysis and metabolic engineering (Metabolic Engineering and Metabolic Flux Analysis). I used the citric acid cycle (labeled with positionally labeled 13C-pyruvate or 13C-alanine) as an example of its utility to probe flux ratios in central carbon metabolism of microorganisms and a simple aquatic C3 plant, Lemna minor (Citric Acid Cycle Labeling In C3 Plants).
My main interest is in plants, and because they normally only use CO2 from the atmosphere as sole carbon source, it is essential to understand what can be gleaned (and equally importantly, what can't be gleaned!) from labeling with 13CO2. We have seen that some 13CO2 is likely to be incorporated into oxaloacetate via the PEP-carboxylase reaction functioning to replenish the citric acid cycle (Citric Acid Cycle Labeling In C3 Plants). This reaction is also of crucial importance in initial CO2 fixation by C4 and CAM plants (C4 Photosynthesis, Crassulacean Acid Metabolism (CAM)). But in all plants ultimately CO2 is mostly fixed by ribulose-1,5-bisphosphate carboxylase (RuBisCO) in the Calvin cycle (The Calvin Cycle of C3 Photosynthesis).
So the questions arise:
How would intermediates of the Calvin cycle become labeled when 13CO2 is supplied to a leaf?
Do all carbon atoms of the Calvin cycle become labeled to the same isotope abundance?
Can we develop simple formulae for calculating the isotopic abundance of Calvin cycle intermediates after supplying 13CO2 of known isotopic abundance.
To address these questions, I developed a computer model of the Calvin cycle (written in Visual Basic) that predicts the time-course of isotopic labeling of each intermediate in the pathway at a specified 13CO2 isotope abundance. The model shown below calculates the time-courses of labeling of intermediates with 0, 1, 2, 3, 4, 5, 6, and 7 labeled atoms in a "pulse" (switch from 1.1% atmospheric 13CO2 to 99% 13CO2) followed by a "chase" (switch back from 99% 13CO2 to 1.1% atmospheric 13CO2 at 2 h):
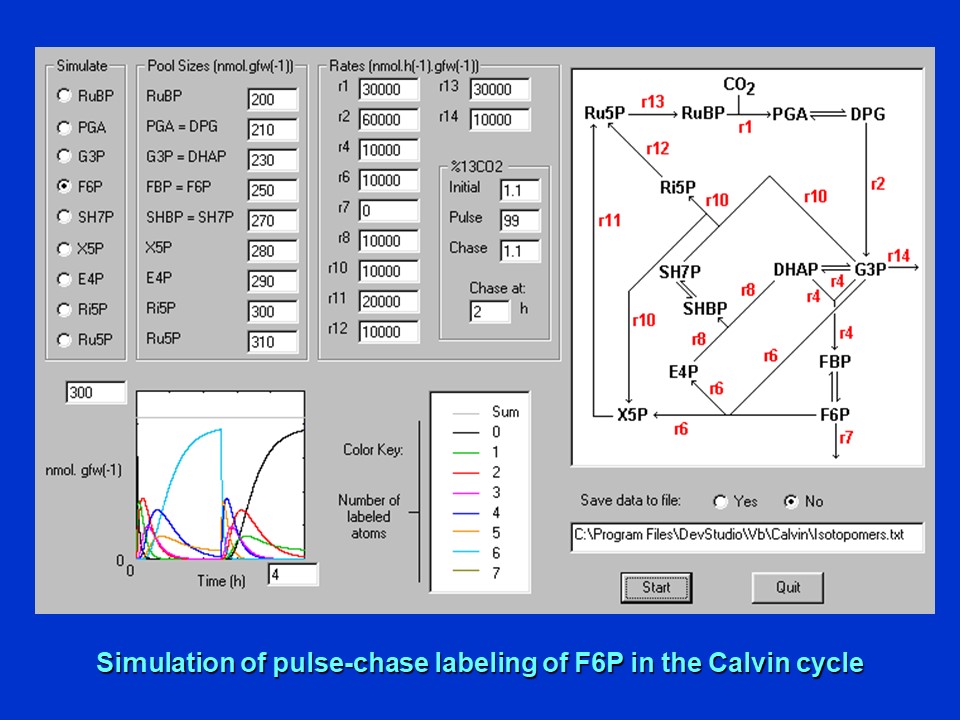
Simulation of the time-course of labeling of fructose-6-phosphate (F6P) in the Calvin cycle during a 2 h pulse with 99% 13CO2 followed by a 2 h chase with 1.1% 13CO2 (D. Rhodes). Please see The Calvin Cycle of C3 Photosynthesis for a detailed explanation of the Calvin cycle intermediates.
Shown in this image is the predicted time-course of labeling of the 6-carbon intermediate, fructose-6-phosphate (F6P) using the assumed fluxes and pool sizes of intermediates specified. This molecule achieves nearly full labeling by 2 h. All 6 carbon atoms of F6P become labeled!
This was found to be the case for all other intermediates of the Calvin cycle (not shown), answering the first 2 questions ... every carbon of every intermediate of the Calvin cycle eventually becomes uniformly labeled with 13C.
To address the 3rd question ... "can we develop simple formulae for calculating the isotopic abundance of Calvin cycle intermediates after supplying 13CO2 of known isotopic abundance?" ... we need to think about probabilities:
For a 3 carbon compound (e.g. 3-PGA) the probability of forming species with 0, 1, 2 or 3 labeled atoms would theoretically be (where C = isotope abundance of 13CO2/100):

For a 4 carbon compound (e.g. E4P) the probability of forming species with 0, 1, 2, 3 or 4 labeled atoms would theoretically be:

For a 5 carbon compound (e.g. RuBP) the probability of forming species with 0, 1, 2, 3, 4, or 5 labeled atoms would theoretically be:
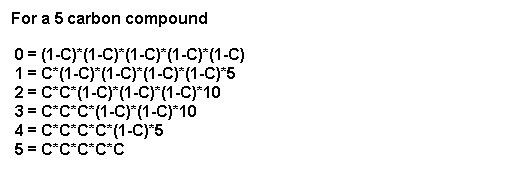
For a 6 carbon compound (e.g. F6P) the probability of forming species with 0, 1, 2, 3, 4, 5, or 6 labeled atoms would theoretically be:

For a 7 carbon compound (e.g. S7P) the probability of forming species with 0, 1, 2, 3, 4, 5, 6 or 7 labeled atoms would theoretically be:

Now, plotting the pool sizes of F6P at the above theoretical isotopic abundances against the model predicted pool sizes of F6P at 4 h (with no chase phase) we see a near perfect correlation between the two methods of calculation:
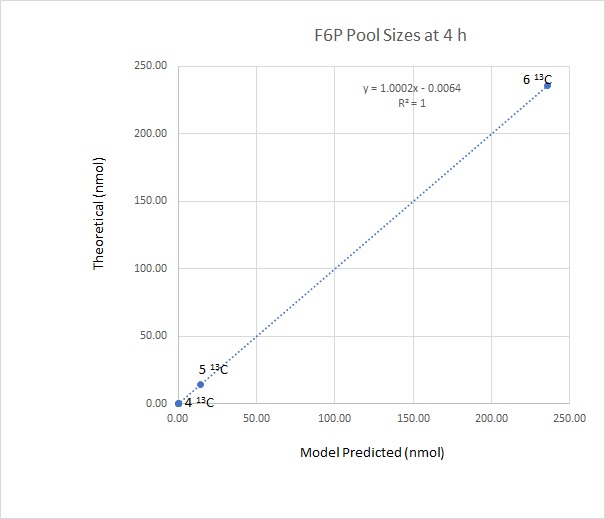
This means that for all intents and purposes, if the pool sizes of Calvin cycle intermediates are small, and the flux through the Calvin cycle is high (as in virtually all plants that are actively photosynthesizing) we can very simply calculate the isotopic abundances of Calvin cycle intermediates from the 13C abundance of the supplied 13CO2.
That's the good news! The bad news is that the operation of the Calvin cycle imparts no positional labeling to any of the intermediates. This means that 13CO2 can't be used like 13C-pyruvate to derive pathway flux information from an isotopic steady-state (Metabolic Engineering and Metabolic Flux Analysis). Rather, in plants the utility of 13CO2 labeling comes from measuring the time-courses of labeling of metabolites derived from Calvin cycle intermediates. This is termed "transient isotopic labeling" (Morgan, J.A., Rhodes, D. Mathematical modeling of plant metabolic pathways. Metab. Eng. 4: 80-89 (2002)).
I will illustrate this in future posts looking at the time-courses of labeling of amino acids in a model C3 plant (Arabidopsis thaliana) and an agriculturally important C4 crop plant (Sorghum bicolor). Stay tuned!
Please feel free to comment or ask questions below. I will attempt to answer as soon as possible.
Thank you for your contribution. Dont forget to link references and sources when applicable!
=======================================================================================
This post was upvoted and resteemed by Steemgridcoin with the aim of promoting discussions surrounding Gridcoin and science.
This service is free. You can learn more on how to help here.
Have a nice day. :)
Yeah, nearly half the dry substance of plants is carbon; and it is conclusively established that they derive, at any rate, the greater part of it, directly from the carbon-dioxide of the atmosphere, which the chlorophyll cells have the power of decomposing in sunlight, at the same time evolving oxygen.
Thumbs up for a well-researched and detailed content sir!