Citric Acid Cycle Labeling In C3 Plants
Introduction
In my previous post on Metabolic Engineering and Metabolic Flux Analysis I described the complex labeling patterns of citric acid cycle intermediates in microbial systems fed with positionally labeled 13C-pyruvate. Much of the complexity arises from the fact that various intermediates of the pathway are withdrawn (e.g. oxaloacetate for aspartate biosynthesis, and alpha-ketoglutarate (2-oxoglutarate) for glutamate synthesis) and, to compensate for this, pyruvate carboxylase functions to anaplerotically replenish intermediates of the pathway.
However, as I mentioned in an earlier post entitled Citric Acid Cycle and Mitochondrial Electron Transport, plants have a different mechanism for replenishing the citric acid cycle. Instead of using pyruvate carboxylase, they employ phosphoenolpyruvate carboxylase. Furthermore, plants often do not operate the complete cycle, using only a portion of it. During nitrogen assimilation for example, plants may only use the part of the cycle up to alpha-ketoglutarate (2-oxoglutarate) and then withdraw that intermediate from the cycle in glutamate biosynthesis (discussed in Citric Acid Cycle and Mitochondrial Electron Transport). As a consequence of these differences, the labeling patterns of the citric acid cycle in C3 plants, when supplied with 13C-pyruvate, are very much simplified.
To illustrate these points I would like to walk you through experiments conducted with the duckweed (Lemna minor) supplied with 13C-alanine labeled in different positions. The work to be described below was briefly reported as part of a book chapter published in 2009 (Amy J.M. Colón, John A. Morgan, Natalia Dudareva, David Rhodes. Application of Dynamic Flux Analysis in Plant Metabolic Networks. In "Plant Metabolic Networks". Springer. pp 285-305 (2009)). Here I would like to offer more detail about how the experiments were conducted, provide a more complete set of results, and a more thorough interpretation of data than was possible in the afore-mentioned article.
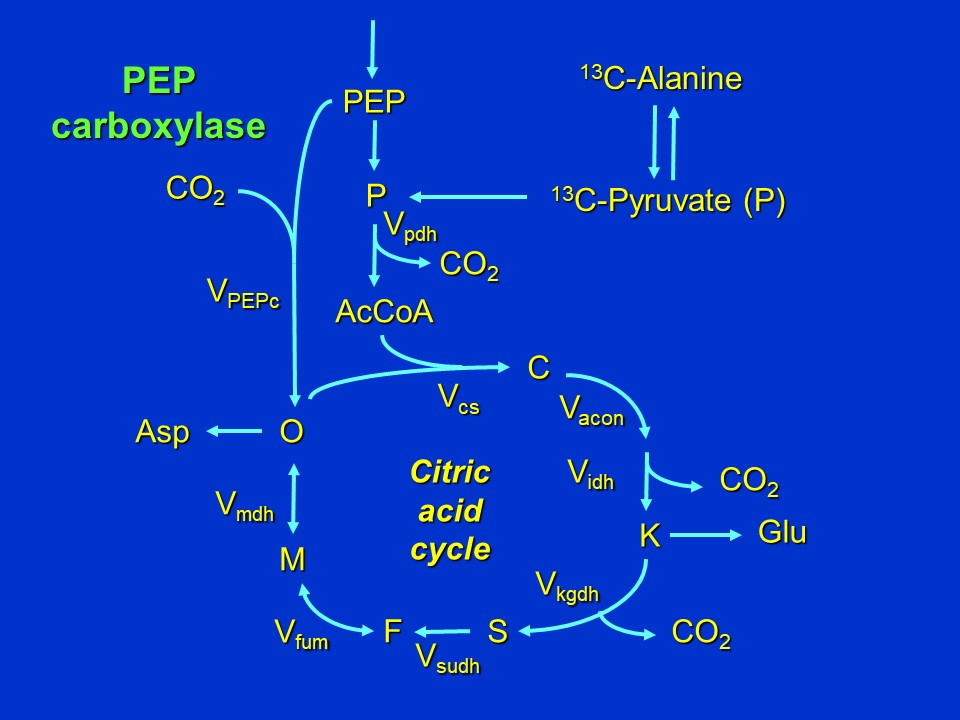
Citric acid cycle when PEP carboxylase is employed for anaplerotic synthesis of oxaloacetate (D. Rhodes)
Abbreviations used: P = pyruvate, AcCoA = acetyl CoA, C = citrate, K = alpha-ketoglutarate (2-oxoglutarate), S = succinate, F = fumarate, M = malate, O = oxaloacetate, Asp = Aspartate, Glu = glutamate, V = reaction velocity (Vsubscripts are abbreviations of enzyme names: cs = citrate synthase, pdh = pyruvate dehydrogenase, acon = aconitase, idh = isocitrate dehydrogenase, kgdh = alpha-ketoglutarate dehydrogenase, sudh = succinate dehydrogenase, fum = fumarase, mdh = malate dehydrogenase, PEPc = phosphoenolpyruvate carboxylase).
Because C3 plants do not express an active pathway for converting pyruvate back to PEP (unlike C4 and CAM plants: see C4 Photosynthesis and Crassulacean Acid Metabolism (CAM)), this means that PEP (derived from C3 photosynthesis, export of triose-phosphates from the chloroplast, and glycolysis (Glycolysis) and entering the citric acid cycle as oxaloacetate, will be unlabeled.
Strategy
Plants, unlike microorganisms, do not readily take up pyruvate supplied exogenously. Pyruvate is also rather unstable and expensive when positionally labeled with 13C. These facts led us to explore the use of 13C-alanine as an alternative precursor. 13C-Alanine is more stable, cheaper and can be more readily taken up by plants. Once taken up it is rapidly used in aminotransferase reactions (alanine aminotransferase/transaminase) to produce pyruvate (see Alanine transaminase):
The resulting pyruvate contains the same positional 13C label as the supplied 13C-alanine.
Lemna minor (duckweed) is one of the smallest angiosperms, growing vegetatively on a simple inorganic growth medium that I have described previously (Common duckweed: an excellent experimental organism and food source), fixing CO2 from the atmosphere by the Calvin cycle (The Calvin Cycle of C3 Photosynthesis). It has one of the fastest growth rates of all angiosperms (Ziegler, P., Adelmann, K., Zimmer, S., Schmidt, C., Appenroth, K.-J. Relative in vitro growth rates of duckweeds (Lemnaceae) – the most rapidly growing higher plants. Plant Biology (Special Issue: Duckweed - Research and Application). Volume 17, Issue Supplement s1, p. 33-41 (2015)), and is ideal for stable isotope tracer studies.
Methods
Lemna minor was pre-grown in the light on medium containing 5 mM KNO3 as sole nitrogen source, and the medium was then supplemented with 2 mM concentrations of alanine labeled with 13C in different positions; either the 1, 2 or 3 positions. Plants were maintained under illuminated conditions throughout.

Illuminated Lemna minor (duckweed) was pre-grown on 5 mM KNO3 and then supplied with 13C-alanine labeled in different positions (D. Rhodes)
At various time intervals (0, 0.5, 1, 1.5, 2, 3 and 6 h) after supplying the labeled alanine, samples of the plants were removed and extracted in methanol. 250 nmol of internal standards (alpha-aminobutyrate and alpha-aminoadipic acid) were added, and methanol extracts were then phase-separated with chloroform and water to remove chlorophyll and lipids, and the upper aqueous phase was dried under a stream of air (Analytical Methods: Amino Acid and Onium Compound Extraction). Dried aqueous-phase extracts were redissolved in water and then subjected to ion exchange chromatography to isolate the neutral + basic amino acids, and acidic amino acids using Dowex-50-H+ and Dowex-1-acetate ion-exchange resins, respectively (Analytical Methods: Amino Acid and Onium Compound Separation by Ion Exchange Chromatography). The purified amino acid fractions were then derivatized to their N(O,S)-heptafluorobutyryl isobutyl esters (Analytical Methods: Amino Acid Derivatization to Heptafluorobutyryl Isobutyl (HFBI) Derivatives) and analyzed by gas chromatogrpahy-mass spectrometry (Analytical Methods: Gas Chromatography-Mass Spectrometry (GC-MS) of HFBI Amino Acid Derivatives).
Shown in the image below is a typical chromatogram of the HFBI-derivatized acidic amino acid fraction of a sample of Lemna at t = 0 h (i.e before supplying 13C-alanine). The major peaks correspond to aspartate (Asp), glutamate (Glu) and the internal standard (IS), alpha-aminoadipic acid (Aad) indicated in red:
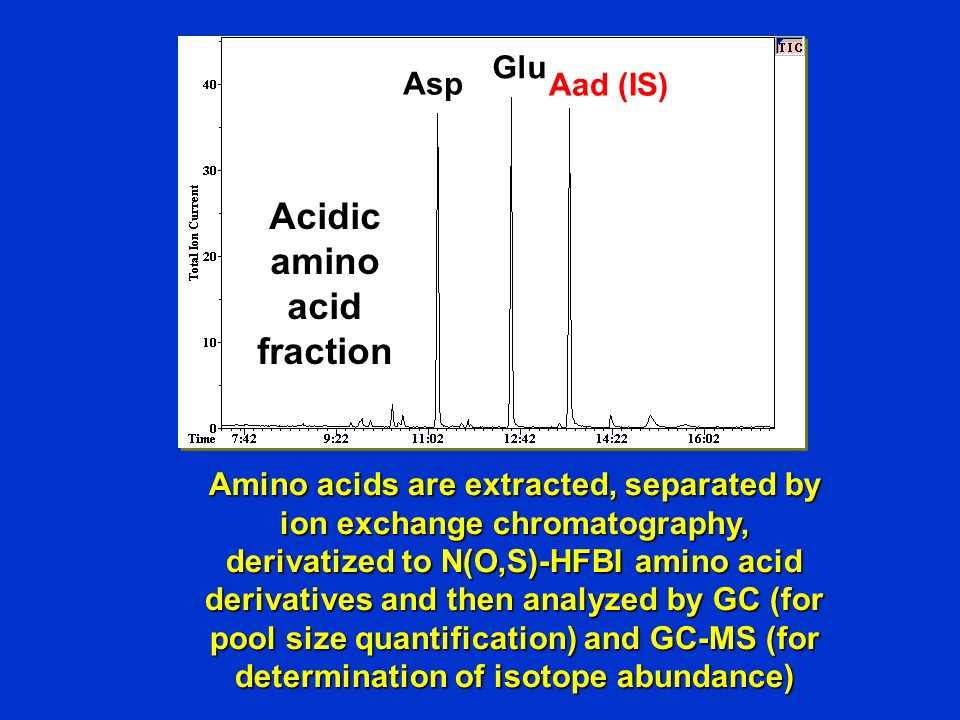
Electron ionization GC-MS chromatogram of HFBI-derivatized acidic amino acid fraction of Lemna at t = 0h (D. Rhodes)
Each peak can be examined for its unique mass spectrum. Shown below is the electron ionization mass spectrum of unlabeled HFBI-glutamate in the acidic amino acid fraction from Lemna at t = 0 h. In this method, 3 major fragment ions of HFBI-glutamate are seen at m/z 298, 280 and 252:
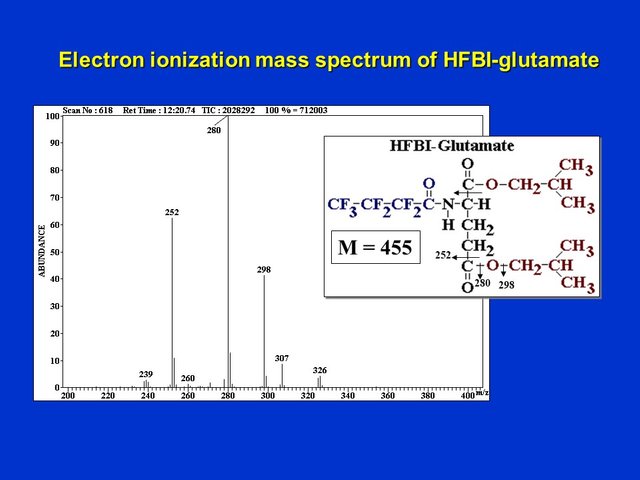
Electron ionization mass spectrum of unlabeled HFBI-glutamate (D. Rhodes)
These fragment ions result first from loss of the isobutyl-esterified alpha-carboxyl (C1) group of glutamate, followed by sequential loss of portions of the isobutyl-derivatized gamma-carboxyl group, as indicated by the arrows. The fragment ions at m/z 298 and 280 both contain the C5 carbon of glutamate, but the fragment at m/z 252 does not! This will become critical in interpreting the positional labeling patterns of glutamate when 13C-alanine is supplied.
Results
Shown below are time-courses of the observed labeling of different fragment ions of glutamate in Lemna supplied with 13C-alanine labeled with a single 13C in either the 1, 2 or 3 positions, together with the expected labeling patterns if alanine (ALA) is converted to pyruvate (PYR), is decarboxylated to acetyl-CoA (AcCoA) and enters the citric acid cycle, generating citrate (CIT), alpha-ketoglutarate (2-oxoglutarate) (KG), and oxaloacetate (OAA), with glutamate (GLU) and aspartate (ASP) being synthesized from alpha-ketoglutarate (2-oxoglutarate) (KG), and oxaloacetate (OAA), respectively. Phosphoenolpyruvate (PEP) is assumed to be the source of the OAA needed to replenish the cycle, with PEP being derived from C3 photosynthesis (PS) via the Calvin cycle, export of triose-phosphates from the chloroplast, and subsequent glycolysis in the cytoplasm.
Note that with 1-13C-alanine as precursor, the isotopic label would be expected to be lost to the atmosphere in the pyruvate decarboxylase reaction at the start of the citric acid cycle, and would therefore not be expected to label acetyl-CoA, citrate, ketoglutarate or glutamate. Consistent with this, the glutamate extracted from plants fed with 1-13C-alanine showed little evidence of isotopic labeling after 3 h (see panels A, B and C which show the percent of glutamate fragment ions containing 0, 1 or 2 13C atoms calculated from the electron ionization mass spectra). Less than 2% isotopic labeling was seen in glutamate using 1-13C-alanine as precursor, and this may have simply been the consequence of partial re-assimilation (via the Calvin cycle (PS)) of the 13CO2 released by pyruvate carboxylase in the citric acid cycle:
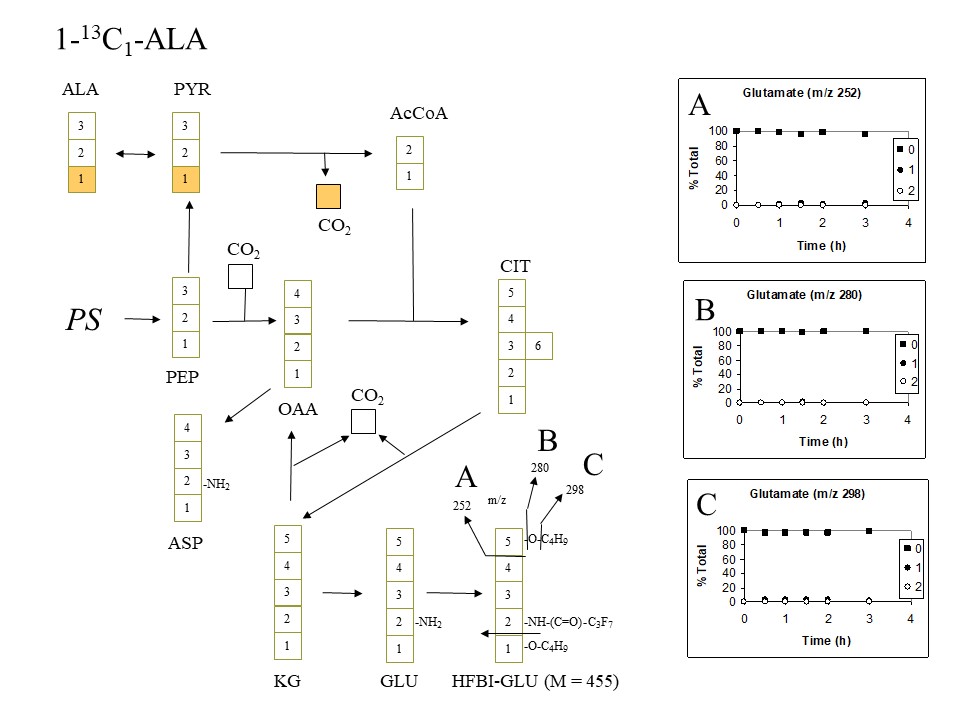
Expected and observed labeling of HFBI-glutamate when 1-13C-alanine is supplied (D. Rhodes)
More convincing evidence for labeling of glutamate with a single 13C-atom in the 5 position is seen when 2-13C-alanine is supplied. The fragment ions of glutamate at m/z 298 and 280 both accumulate a single 13C during the 3 h time-course as the unlabeled glutamate (i.e. glutamate containing no 13C atoms) correspondingly declines as a percent of total glutamate. Notably, the fragment ion of glutamate at m/z 252 contains no 13C (see panel A), as expected if the single 13C incorporated into glutamate via the citric acid cycle is located in the 5 position:
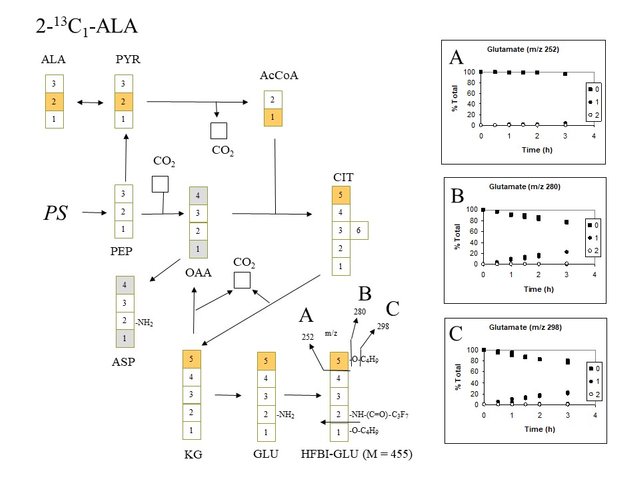
Expected and observed labeling of HFBI-glutamate when 2-13C-alanine is supplied (D. Rhodes)
If alpha-ketoglutarate (2-oxoglutarate) (KG) was not only used in glutamate synthesis, but also participated in a full citric acid cycle it would be expected to contribute label to oxaloacetate (OAA) in the 1 and 4 positions, which would then proceed to label aspartate (ASP) in the same positions (see gray boxes) and in turn label citrate (CIT), KG and GLU with additional 13C. No evidence is seen for substantial labeling of glutamate with more than 1 labeled atom in the 5 position. Therefore, one must conclude that there is very little utilization of KG in a full citric acid cycle.
When 3-13C-alanine is supplied one would expect glutamate to become labeled in the C4 position, and indeed that is what is observed. All three fragment ions of glutamate (m/z 252, 280 and 298) acquire a single 13C atom:
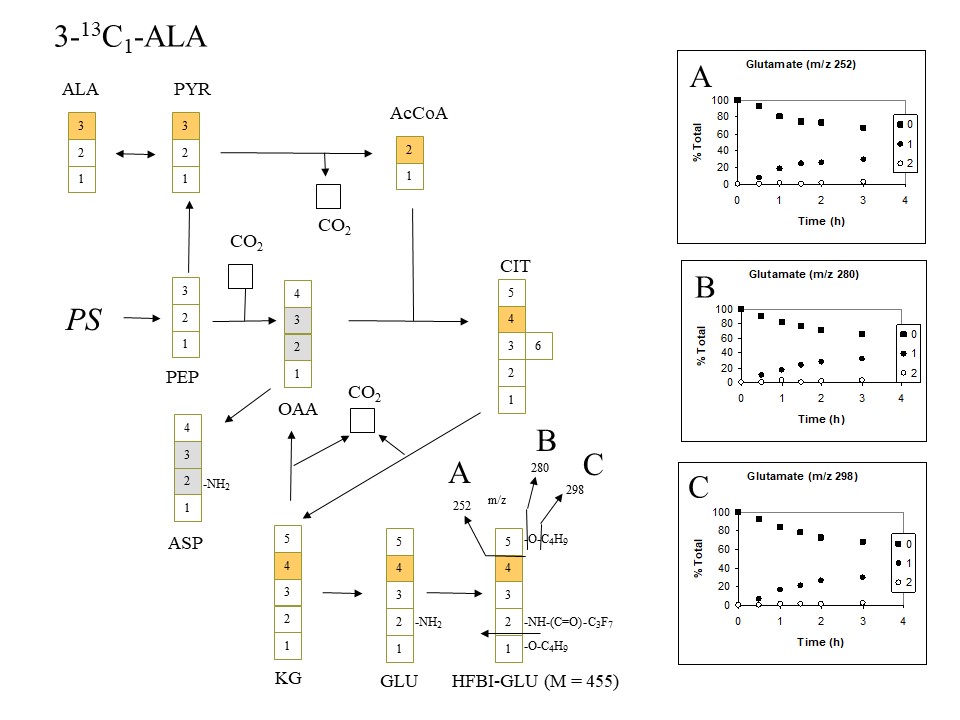
Expected and observed labeling of HFBI-glutamate when 3-13C-alanine is supplied (D. Rhodes)
If alpha-ketoglutarate (2-oxoglutarate) (KG) was not only used in glutamate synthesis, but also participated in a full citric acid cycle it would be expected to contribute label to oxaloacetate (OAA) in the 2 and 3 positions, which would then proceed to label aspartate (ASP) in the same positions (see gray boxes) and in turn label citrate (CIT), KG and GLU with additional 13C. No evidence is seen for substantial labeling of glutamate with more than 1 labeled atom in the 4 position. Again, one must conclude that there is very little utilization of KG in a full citric acid cycle.
This was confirmed by analyzing the labeling patterns of alanine, aspartate, glutamate and glutamine during a 6 h time-course of labeling with 3-13C-alanine. Alanine became rapidly labeled (as a result of uptake from the growth medium) to about 60% within the first 1.5 h. Glutamate and glutamine both became labeled with a single 13C atom to about 35 to 38% after 6 h, while aspartate became labeled with a single 13C atom to only about 3 -4 % after 6 h. This suggests that the citric acid cycle in illuminated Lemna grown on nitrate as nitrogen source is mostly operating as a partial cycle as discussed previously (see also Citric Acid Cycle and Mitochondrial Electron Transport):
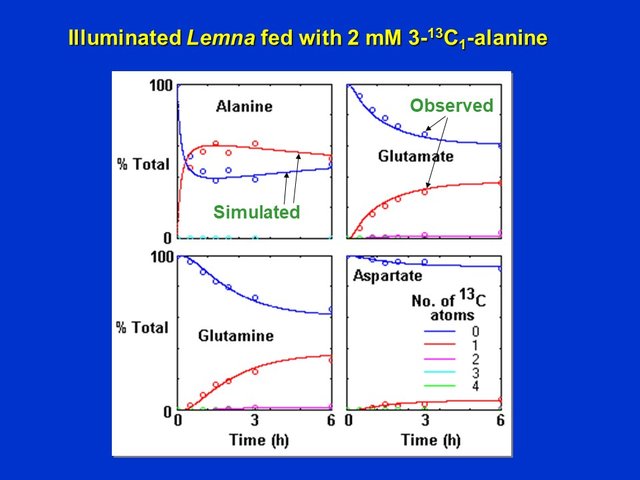
Time-course of labeling of alanine, glutamate, gutamine and aspartate in Lemna supplied with 2 mM 3-13C-alanine for up to 6 h. Amino acid species with 0, 1, 2, 3 and 4 labeled atoms were quantified by analyses of the mass spectra of the HFBI-derivatized amino acids in the neutral + basic (alanine and glutamine) and acidic (glutamate and aspartate) amino acid fractions separated by ion exchange chromatography (D. Rhodes)
To further confirm these findings, we then fed uniformly (U) labeled 13C-alanine (i.e. alanine labeled in all 3 positions) to illuminated Lemna for 8 h.
Unlabeled alanine normally shows a mass spectrum with major ions at m/z 342 (the unfragmented molecular (M+H+) ion of HFBI-alanine), and a fragment ion (M-101) at m/z 240, resulting from loss of the alpha-carboxyl (C1) group of HFBI-alanine. Consistent with this, the mass spectrum of the intracellular alanine pool of Lemna fed with U-13C-alanine for 8 h showed a mass shift of 3 atomic mass units (amu) for the molecular ion (343 --> 345), and a mass shift of only 2 amu for the fragment ion missing C1 of alanine (240 --> 242):
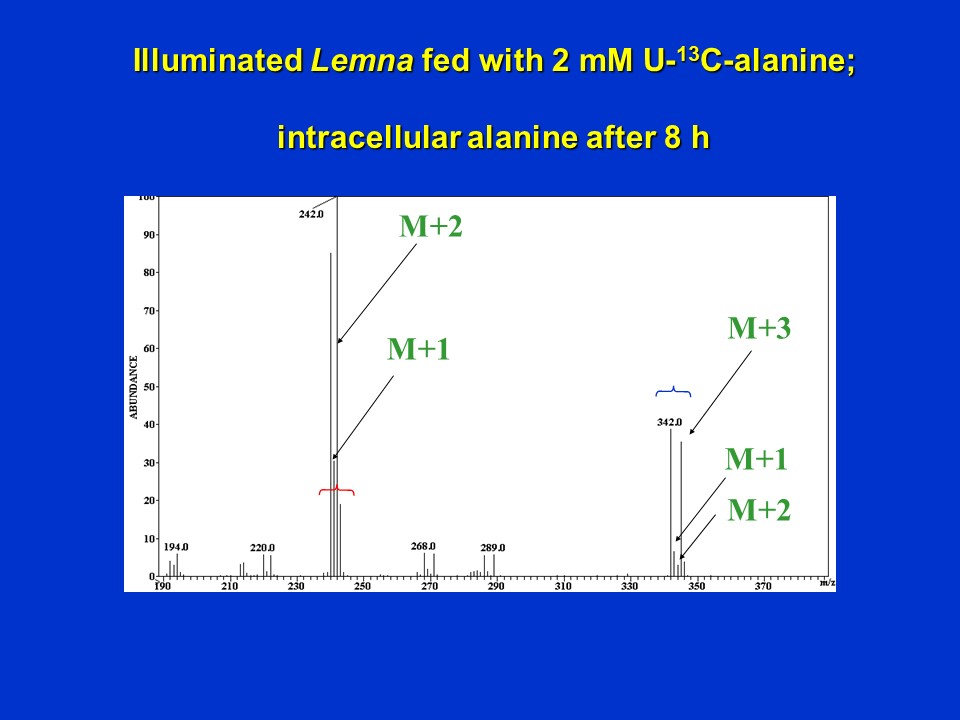
Electron ionization mass spectrum of intracellular alanine (derivatized to HFBI-alanine) extracted from Lemna supplied with 2 mM U-13C-alanine for 8 h (D. Rhodes)
As noted earlier, unlabeled HFBI-glutamate shows a mass spectrum with major ions at m/z 298, 280 and 252. The molecular (unfragmented) (M+H+) ion of unlabeled glutamate occurs at m/z 456. As expected for operation of the partial citric acid cycle the molecular (unfragmented) (M+H+) ion of HFBI-glutamate acquires only 2 labeled atoms (note the mass shift from 456 --> 458), the fragment ions at m/z 298 and 280 acquire predominantly 2 labeled atoms (298 --> 300, 280 ---> 282), and the fragment ion at m/z 252 acquires predominantly a single 13C (252 --> 253):
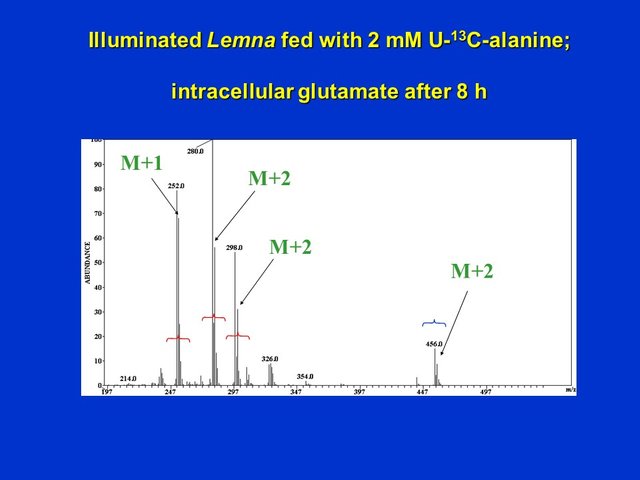
Electron ionization mass spectrum of intracellular glutamate (derivatized to HFBI-glutamate) extracted from Lemna supplied with 2 mM U-13C-alanine for 8 h (D. Rhodes)
Operation of the full citric acid cycle would have been expected to label glutamate in more positions than observed!
Conclusions
13C-labeled alanine is a good surrogate for pyruvate in order to label the citric acid cycle in plants. It is cheaper than pyruvate, more stable, and more readily absorbed by plant tissues than pyruvate.
The results described above are consistent with the operation of a partial citric acid cycle during nitrogen assimilation in the C3 plant Lemna minor. PEP-carboxylase provides the anaplerotic pathway of synthesis of oxaloacetate required to sustain this pathway when alpha-ketoglutarate (2-oxogluratate) and oxaloactete are removed for glutamate and aspartate synthesis, respectively. Very little of the alpha-ketoglutarate (2-oxogluratate) produced is processed back to oxaloacetate (via succinate, fumarate and malate) via the complete citric acid cycle (Sweetlove, L.J., Beard, K.F., Nunes-Nesi, A., Fernie, A.R., Ratcliffe, R.G. Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci. 15: 462-470 (2010)). With operation of the complete citric acid cycle much heavier labeling of aspartate would have been expected, and glutamate should have received label in more than just the C4 and C5 positions.
Please note that these conclusions are valid only for C3 plants which lack a mechanism for rapid conversion of PEP to pyruvate (PYR). C4 plants and CAM plants have evolved mechanisms which employ PEP-carboxylase to concentrate CO2, and enzymatic machinery for converting PEP back to PYR (see C4 Photosynthesis and Crassulacean Acid Metabolism (CAM)). The labeling patterns of glutamate and aspartate might be expected to be significantly different in C4 and CAM species!
Please feel free to comment or ask questions below. I will attempt to answer as soon as possible.
Great and descriptive post. Well explained. I felt like I was in an actual plant physiology lecture.
Great article! Bookmarked for future reference. How does CO2 (or HCO3-) get incorporated in bacteria?
Photosynthetic cyanobacteria can fix CO2 (or HCO3-) like plants (Calvin cycle and PEP-carboxylase). But these organisms are unlikely to be of relevance in a cave environment with no light!
Many bacteria can trap CO2 (or HCO3-) via pyruvate carboxylase and incorporate it into citrate, oxaloacetate, and aspartate (see earlier post on Metabolic Engineering and Metabolic Flux Analysis).
But some bacteria, and I think these would be the most interesting in a cave environment, metabolizing CaCO3, would be capable of running the citric acid cycle (TCA cycle or Krebs Cycle) in reverse (see Reverse Krebs cycle). Recently a novel variant of this reverse TCA cycle has been discovered:
Nunoura, T., Chikaraishi, Y., Izaki, R., Suwa, T., Sato, T., Harada, T., Mori, K., Kato, Y., Miyazaki, M., Shimamura, S., Yanagawa, K., Shuto, A., Ohkouchi, N., Fujita, N., Takaki, Y., Atomi, H., Takai, K. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science 359: 559-563 (2018)
I'll check that out. We found lots of Nitrospira and others capable of using nitrite/HCO3 (chemolithotrophs) and Fe/Mn sulphur bacteria.
Nitrospira seems to use a reverse TCA cycle for CO2 fixation:
Lücker, S., Wagner, M., Maixner, F., Pelletier, E., Koch, H., Vacherie, B., Rattei, T., Damsté, J.S., Spieck, E., Le Paslier, D., Daims, H. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U.S.A. 107: 13479-13484 (2010)
Another good one! This was the info we needed at the same time we were finding and hypothesizing. My expertise was in technical methodology and experimental approaches to get the data and compile sequences. Colleagues were better than me at interpretation of the physiological systems. Some of my microbiology collaborators include Penelope Boston, Hazel Barton, Diana Northup. Recently I discovered that many phylogenetic trees from artificial substrates were not published in a recent book, so they are fair game for a new post!
I've read that one several times now. I last worked on that in 2007-2008, and I had no idea so much was known! From 2008-2011 I was looking at Bacteroides 16S sequences. I'm working on some of our Mammoth Cave bacterial data.
The pace of discovery in biological sciences is staggering ... it's getting hard to keep up with the literature.