Is Bread Certain Death?
Summary
This post represents a response to the suggestion of @lesshorrible in his post NSAS Idea Suggestion #14 - Is Bread Certain Death?
Bread. Image Source:
The short answer to this question is ... NO, but it can make certain individuals very sick.
About 0.5 to 1.0% of the human population are affected by a disease called celiac disease (also called coeliac disease, or celiac sprue, or gluten-sensitive enteropathy) caused by a sensitivity to proteins (gluten) in wheat (and other cereals). Gluten ingestion (bread intake) triggers an autoimmune cascade leading to destruction of villi of the small intestine, abdominal pain, diarrhea, and impaired absorption of vitamins and minerals. These individuals benefit from a strict gluten-free diet. Celiac disease is one of the best studied autoimmune diseases in humans and has provided a great deal of insight concerning the link between environment (in this case bread intake) and our genes in triggering autoimmunity.
If untreated, celiac disease may result in cancers such as intestinal lymphoma and a slightly increased risk of early death (Holmes and Muirhead (2018)). However, celiac disease can become potentially deadly if it is combined with a liver disease called primary sclerosing cholangitis (PSC), which shares many of the genetic risk factors with celiac disease, as will be discussed in detail below (Grainge et al. (2011)). PSC is an autoimmune liver disease of unknown etiology that is often associated with inflammatory bowel disease (mostly ulcerative colitis, and sometimes Crohn's disease) and has a male predominance. PSC carries a tremendously high risk of liver failure, colorectal cancer and cholangiocarcinoma (LaRusso et al. (2006)). Gluten withdrawal has no effect on the progress of PSC. Deaths in celiac disease are more prevalent in males than in females despite the fact that celiac disease is more prevalent in females than in males (Grainge et al. (2011)). But death is not certain!
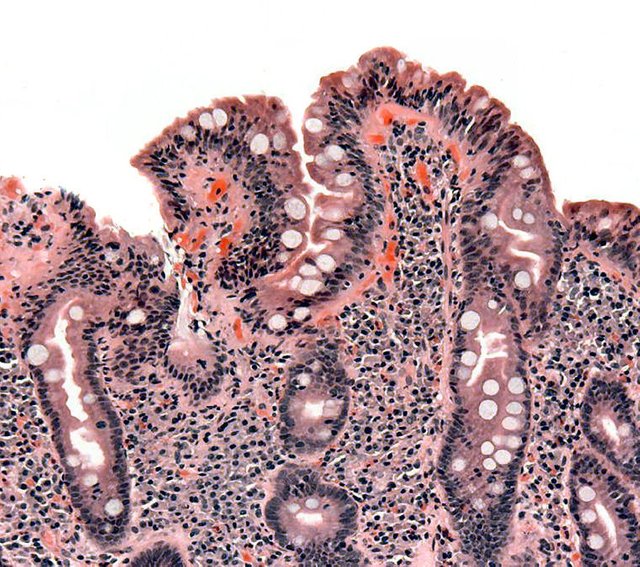
Biopsy of small bowel showing coeliac disease manifested by blunting of villi, crypt hyperplasia, and lymphocyte infiltration of crypts, consistent with Marsh classification III. Released into public domain on permission of patient. Image Source:
A brief note on autoimmune diseases
Most autoimmune diseases are thought to be initiated by an environmental trigger in genetically susceptible individuals. For some autoimmune diseases the environmental triggers may be infectious agents; e.g. bacteria or viruses. In others, the triggers may be environmental toxins, or proteins found in the food supply. Deficiencies of certain vitamins (e.g. vitamin D) and smoking are commonly recognized environmental factors which may also influence susceptibility. There is increasing evidence that the microbiota of the gut may play a critical role in disease initiation and progression. The disease process can often lead to the production of autoantibodies; these are sometimes useful for diagnosis, and in many cases are intimately involved in the pathogenesis of the disease. Often males and females will show different susceptibilities, and so a gender effect is frequently recognized, with females being more susceptible for most (but not all) autoimmune diseases.
Overview of celiac disease
Celiac disease (also called coeliac disease, or celiac sprue, or gluten-sensitive enteropathy) is probably the best understood of the autoimmune diseases. It is caused by a sensitivity to the proteins (gliadins and glutenins (gluten proteins)) in certain grass seeds; primarily wheat, barley, and rye. As wheat is the major ingredient in bread, celiac disease is primarily associated with bread intake, although many other food products include wheat, and other cereals as ingredients. Thus, the environmental 'trigger' for this disease is well characterized: peptides (short chains of amino acids) derived from the breakdown of gluten. The most effective treatment for this disease is maintenance of a gluten-free diet. Celiac disease is remarkably common, and is estimated to affect 0.5 to 1% of the population of Europe and the United States (Megiorni and Pizzuti (2012)). Nelson (2002) estimates that about 1 in every 250 persons in the USA have celiac disease, but many may be undiagnosed because the disease is often "silent". The female:male ratio is about 2:1 (Megiorni and Pizzuti (2012)).
Celiac disease is generally characterized by an atrophy of the villi of the small intestine and can be accompanied by diarrhea, fatty stools, abdominal pain and distention, weight loss, chronic fatigue, migraines, depression, osteoporosis/osteopenia and infertility (Taylor et al. (2015)). The disease can result in malabsorption of nutrients, such as vitamins, folates, iron, zinc and calcium. Moreover, a large number of extraintestinal manifestations of the disease can also occur, including dermatitis herpetiformis (a skin rash), arthritis, neuropathy, and liver disease. Celiac disease patients seem to be at higher risk of other autoimmune diseases, including type 1 diabetes, multiple sclerosis, psoriasis, primary biliary cirrhosis/cholangitis, inflammatory bowel diseases, autoimmune hepatitis, autoimmune thyroid disorders, and primary sclerosing cholangitis (Ch'ng et al. (2007), Assa et al. (2017); Lodhi et al. (2018), Narciso-Schiavon and Schiavon (2017), Marciano et al. (2016)). Celiac disease is usually diagnosed by serum antibody tests (see below), an intestinal biopsy, and mitigation of symptoms following gluten withdrawal.
Antibodies
Celiac disease is characterized by antibodies to a protein called transglutaminase 2 (TG2). The enzyme transglutaminase 2 not only converts the gluten-derived peptides into forms that make them more immunogenic, but it also cross-links the immunogenic peptides with itself and other proteins. These cross-linked proteins appear to become the targets for auto-antibody production by B cells. TG2 antibody production in celiac disease is extensively discussed by De Re et al. (2017). Some patients also devlop anti-endomysium (EMA) auto-antibodies (Megiorni and Pizzuti (2012)).
Genetic susceptibility caused by genes in the major histocompatibility complex
The genetic susceptibility to celiac disease is well characterized, but highly complex. One of the most important genetic determinants of susceptibility is a pair of genes in the major histocompatibility complex (MHC) in the human leukocyte antigen (HLA) class II region on the short arm of human chromosome 6. The pair of adjacent genes involved are the HLA-DQA1 and HLA-DQB1 genes. Certain variants (alleles) of these two genes are known to confer a high risk of celiac disease (CD). "About 90-95% of CD patients carry DQ2.5 heterodimers, encoded by DQA1-05 and DQB1-02 alleles both in cis or in trans configuration, and DQ8 molecules, encoded by DQB1-03:02 generally in combination with DQA1-03 variant." (Megiorni and Pizzuti (2012)).
It is noteworthy that this same genetic region (although not necessarily the exact same gene variants/alleles!) seems also to be important in a number of other autoimmune diseases. These include autoimmune hepatitis, alopecia aerata, myasthenia gravis, autoimmune polyglandular syndrome, antiphospholipid syndrome, primary biliary cholangitis/cirrhosis, psoriasis, psoriatic arthritis, ankylosing spondylitis, primary sclerosing cholangitis, ulcerative colitis, Crohn’s disease, multiple sclerosis, type 1 diabetes, sarcoidosis, Sjogren’s syndrome, systemic lupus erythrematosus, and autoimmune thyroid disease. I propose to address each of these autoimmune diseases in future articles.
It is important to emphasize that if you carry the above HLA genetic risk factors for celiac disease, it does not necessarily mean that you will develop the disease in your life-time. "Nevertheless, it is an important tool able to discriminate individuals genetically susceptible to celiac disease, especially in at-risk groups such as first-degree relatives (parents, siblings and offspring) of patients and in presence of autoimmune conditions (type 1 diabetes, thyroiditis, multiple sclerosis) or specific genetic disorders." (Megiorni and Pizzuti (2012)).
On a personal note, I have submitted by own DNA to 23andMe and I have received this report:
"We detected a variant linked to the HLA-DQ2.5 haplotype".
"You have a slightly increased risk of developing celiac disease based on your genetic result. However, most people with this result do not develop celiac disease. Consider discussing your risk with a healthcare professional, especially if you have a family history or other risk factors for this condition."
"People with this result have a slightly increased risk of developing celiac disease. However, studies estimate that only about 3% of people with one or more copies of the HLA-DQ2.5 or HLA-DQ8 haplotypes develop celiac disease."
As noted above, two of the major genetic determinants for celiac disease are the alleles HLA-DQA1-05 and HLA-DQB1-02 (Medrano et al. (2012)). These alleles are carried on the so-called 8.1 ancestral haplotype (AH8.1) which is remarkably conserved in the Northern European population (Karlsen et al. (2007)). The name AH8.1 refers to the alleles B8, A1 in the large block of genes in the major histocompatibility complex HLA A1-B8-DR3-DQ2. This haplotype seems to be unusually resistant to recombination. It may have originated about 20,000 years within Europe. This haplotype is also strongly associated with susceptibility to primary sclerosing cholangitis (PSC) (Karlsen et al. (2007)).
The HLA-DQA1 and HLA-DQB1 genes encode proteins that are expressed on the surface of antigen presenting cells and which pair up with each other: i.e. they form heterodimers (meaning dimers formed from two distinct proteins). The specific variants (alleles) of these 2 genes allow gluten to become toxic because the dimers formed from these two proteins bind specifically to certain peptides formed when gluten is partially digested. These gluten peptides are rich in the amino acids glutamine and proline. Transglutaminase 2 acts on these peptides, and converts the glutamine residues to glutamates. This gives the peptides a special ability to bind to the unique HLA-DQA1/HLA-DQB1 heterodimers found in celiac disease patients. The heterodimers bound to gluten peptides are then presented by antigen presenting cells to T cells, resulting in a complex inflammatory cascade (release of inflammatory cytokines) and tissue damage. The possibility that there may be other genes within the MHC/HLA region influencing susceptibility to celiac disease cannot yet be eliminated (Medrano et al. (2012)).
Non-HLA genetic risk factors for celiac disease
Not all of the pieces of the celiac disease puzzle are worked out yet, but it is known that the gene CTLA-4 (cytotoxic T lymphocyte associated) (located on chromosome 2q33) may also contribute to celiac disease susceptibility. The CTLA-4 gene produces a protein that normally acts as a negative regulator of T cell activation. Mutations in the CTLA-4 gene can result in uncontrolled T cell activation, and therefore increased susceptibility to a number of autoimmune diseases. Likewise, PTPN22 plays a key role in regulating T cell activation and effector responses and is associated with a number of autoimmune diseases, including celiac disease (Aflatounian et al. (2017)).
Interleukin 2 (IL2) and interleukin 21 (IL21) play essential roles in the immune system, via direct effects on T cells. Polymorphisms in the chromosomal region encoding IL2 and IL21 have been associated with celiac disease (Guo et al. (2015)) and primary sclerosing cholangitis (Stallhofer et al. (2011)).
SH2B3 encodes a protein called LNK that functions as a regulator in signaling pathways relating to hematopoiesis, inflammation, and cell migration. Polymorphisms in the SH2B3 gene have been associated with celiac disease (Guo et al. (2015)) and primary sclerosing cholangitis (Webb and Hirschfield (2016)).
A genetic variant in the myosin IXB (MYO9B) gene (chromosome 19p13.11) has been associated with celiac disease (Koskinen et al. (2008)). This gene may cause an impairment of the intestinal barrier, which may explain why immunogenic gluten peptides are able to pass through the epithelial barrier. This gene represents an interesting candidate for the IBD6 gene, also located on chromosome 19p13, which confers susceptibility to both Crohn’s disease and ulcerative colitis.
An additional gene determining susceptibility to celiac disease is the MBL2 (mannose binding lectin) gene located on chromosome 10q11.2-q21 (Boniotto et al (2005)). This particular gene is also associated with Crohn’s disease (Choteau et al. (2016b)) and plays a critical role in fungal (Candida albicans) elimination from the gut (Choteau et al. (2016a)).
A gene on chromosome 11q has been implicated in susceptibility to celiac disease. This gene may represent the matrix metalloproteinase 3 gene (MMP-3) (stromelysin-1) (chromosome location 11q23) (Mora et al. (2005)). Variants of this gene appear to be a risk factor for celiac disease only in men; this is different from the HLA susceptibility alleles that confer a higher risk in women. The MMP-3 gene has also been associated with PSC susceptibility and disease progression (Juran et al, (2011)). The considerable overlap between the genes conferring susceptibility to celiac disease, and those conferring susceptibility to PSC and/or inflammatory bowel diseases described above, may explain why these diseases can often co-exist.
The following image (created by myself) summarizes the partial sequence of events, articulated above, leading to celiac disease:
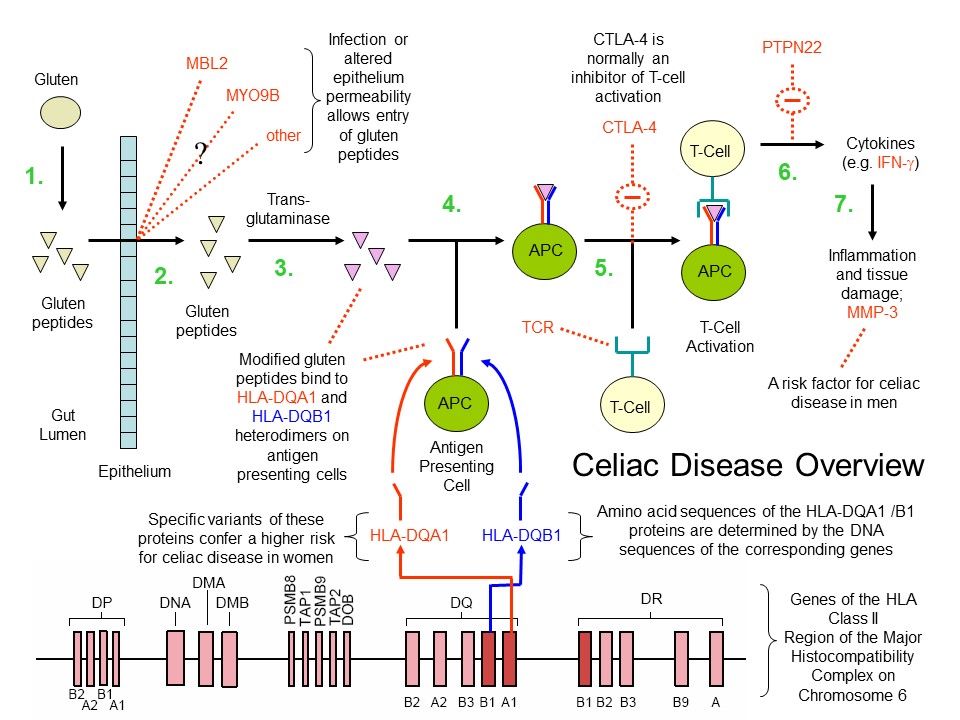
Celiac disease overview (D. Rhodes). 1. Gluten protein is ingested and broken down into small peptides in the gut. 2. Gluten peptides are absorbed. Various celiac disease susceptibility genes may influence gut barrier permeability/integrity. 3. Gluten peptides are modified by trans-glutaminase. 4. Antigen presenting cells carrying HLA-DQA1 and HLA-DQB1 heterodimers bind the modified gluten peptides. 5. and 6. Antigens are presented to T cells leading to their activation and release of pro-inflammatory cytokines such as IFN-gamma. Various celiac susceptibility genes (e.g. CLTA-4, PTPN22, IL2, IL21) may influence T cell activation and migration (not shown). Genes involved in T cell migration and their tissue homing may include CCR3 (Romanos et al. (2009)) and ITGA4 (Garner et al. (2009)). 7. Inflammation and tissue damage ensues. Inflammation may be modulated by certain gene (e.g. MMP-3). Not depicted in this sequence (for simplicity) is the further involvement of T cells in promoting the production of B cells that generate autoantibodies to trans-glutaminase 2.
FUT2 (encoding Fucosyltransferase 2) is a susceptibility gene for celiac disease (Parmar et al. (2012a)), PSC and Crohn's disease (Maroni et al. (2015)). In PSC it determines risk of biliary infection with Candida albicans (Rupp et al. (2014)). FUT2 is implicated in regulating the gut microbiome composition (Wacklin et al. (2011), Rausch et al. (2011)) and has a dramatic effect on susceptibility to norovirus infection, a leading cause of acute gastroenteritis (Currier et al. (2015)).
These genetic findings have spurred interest in examining whether the gut microbiome, fungal pathogens and viruses may have an influence on celiac disease (De Re et al. (2017), Olivares et al. (2018), Sebastián Domingo et al. (2018), Cukrowska et al. (2017)). Bouziat et al. (2017) have proposed that reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease (see also: Verdu and Caminero (2017)). Intriguingly reovirus, normally an innocuous virus, is capable of suppressing anti-inflammatory regulatory T cells, and promoting pro-inflammatory Th1 cells in response to dietary antigen (Bouziat et al. (2017)).
Genes conferring susceptibility to celiac disease continue to be identified (see e.g. FRMD4B (Garner et al. (2014); IL12A, KIAA1109, SCHIP1 (Plaza-Izurieta et al. (2011); BACH2, CCR4, CD80, RUNX3 (Dubois et al. (2010)) but remain to be fully integrated into the celiac disease puzzle. Certainly many of these genes influence T cell biology and immune cell function (Dubois et al. (2010)).
The "Online Mendelian Inheritance in Man" website (OMIM) lists the following Celiac disease susceptibility loci:
CELIAC1 (6p21.32), associated with the closely linked HLA-DQA1 and HLA-DQB1 pair of genes described above.
CELIAC2 (5q31-q33) gene not yet identified. The latter gene does not appear to be the same as the IBD5 gene mapping to this same region.
CELIAC3 (2q33.2), associated with CTLA4.
CELIAC4 (19p13.11), associated with myosin IXB (MYO9B).
CELIAC5 (15q11-q13), gene not yet identified.
CELIAC6 (4q27), associated with IL2 and/or IL21 genes (Romanos et al. (2009)).
CELIAC7 (1q31), associated with RGS1 (Romanos et al. (2009)).
CELIAC8 (2q11-q12), associated with the IL18RAP and IL18R1 (Romanos et al. (2009)).
CELIAC9 (3p21), associated with a cluster of chemokine receptor genes. CCR3 is a likely candidate (Romanos et al. (2009)).
CELIAC10 (3q25-q26), associated with the IL12A gene (Romanos et al. (2009)).
CELIAC11 (3q28), near the LPP gene (Romanos et al. (2009), Garner et al. (2009))
CELIAC12 (6q25.3), near the TAGAP gene (Romanos et al. (2009)).
CELIAC13 (12q24), associated with the SH2B3 gene (Romanos et al. (2009)).
Non-celiac gluten or wheat sensitivity
A new disease called non-gluten (or wheat) sensitivity (NCGS) has recently been recognized (Kabbani et al. (2014), Bathrellou et al. (2018)). It's diagnosis relies on exclusion of wheat allergy and celiac disease (antibodies, genetic (HLA) DQ2/DQ8 testing, biopsies) and improvement of symptoms on a gluten free diet (Mansueto et al. (2014)). It appears that NCGS patients may be a quite heterogenous group, and, thus far, no reliable biomarkers for the different potential sub-groups have been identified (Mansueto et al. (2014)).
Modern wheat
A recent article at Grainstorm Heritage Baking, entitled What's wrong with modern wheat, has pointed out that modern-day processing of wheat is radically different from how it used to be performed. Many of the nutrients of wheat are literally stripped out of wheat flour during the modern-day milling process. To what extent the emergence of the new disease entity "Non-celiac gluten or wheat sensitivity" is driven by today's processed, nutrient poor wheat flour has not yet been determined. Is the modern-day "gluten-free bandwagon" in part driven by misinformation, and in part by "something wrong with modern wheat"? (What's wrong with modern wheat). Let me know what you think about this is the comments section below.
For discussion of the importance of wheat as a source of calories and nutrients in human health I can recommend the recent articles written by Peter Shewry (Shewry et al. (2015), Shewry and Hey (2016)). Wheat provides .... "protein, vitamins (notably B vitamins), dietary fiber, and phytochemicals. Of these, wheat is a particularly important source of dietary fiber, with bread alone providing 20% of the daily intake in the UK, and well-established relationships between the consumption of cereal dietary fiber and reduced risk of cardio-vascular disease, type 2 diabetes, and forms of cancer (notably colo-rectal cancer)." (Shewry and Hey (2016)). Wheat is an excellent source of glycine betaine (betaine), a key nutrient for human health that is capable of lowering blood homocysteine levels and compensating for folate or vitamin B12 deficiencies (Ross et al. (2014)).
Modern varieties of wheat are not significantly different from older varieties in the levels of the gliadins that trigger coeliac disease in susceptible individuals (De Santis et al. (2017)). Moreover the type of gliadins that are responsible for wheat allergy (including wheat dependent exercise induced anaphylaxis (WDEIA)) have been drastically reduced by modern breeding practices (De Santis et al. (2017)).
Wheat domestication was a major event in the establishment of human civilization (Zou et al. (2015)). Wheat was among the original plant species to be domesticated by humans (the dawn of agriculture). It was first cultured by humans in the Fertile Crescent about 11,000 - 10,000 years ago, shortly after the end of the Younger Dryas. Despite the modern "gluten-free" trend of today, wheat is still considered as one of the most important crops in the world (Zou et al. (2015))!
Additional Resources
- Celiac Disease Foundation
- Celiac Sprue Association
- Celiac Disease - Medline
- Celiac disease - PubMed
- Coeliac Disease - Wikipedia
- HLA-DQ - Wikipedia
- HLA A1-B8-DR3-DQ2
- Wheat - Wikipedia
References
Verdu, E.F., Caminero, A. How infection can incite sensitivity to food. Science 356: 29-30 (2017)
When we don't understand something, we always accuse environmental and genetic factors, medically speaking any kind of sensitivity may kill you in extreme cases like anaphylactic shocks or ionic disturbances due to chronic diarrehea but saying bread can kill you is a little bite exaggerating, anything in excess or deficiency is harmeful. I personally blame the poor life style and the poor diets, the average sugar intake increased exponentially last years. I read a study last month on the effects of salt intake over 16 years, they found that low-salt diets increase the cardiovascular risk just like the high diets.The point here is we lack critical knowledge to understand the whole phenomenon,, anyway I really enjoyed your post, great effort !
Agreed. I didn't like the title much, but had to use it to address the @nsas question posed by @lesshorrible.
Yep, that title is my fault, sorry. You do not have to use the proposed titles btw. I should have mentioned that. I love what you make out of the suggested ideas though! Cheers!
No problem. It gave me an incentive to try to argue against the idea. Thanks again for letting me address this issue. Cheers.
Yeah, it was meant to be sarcastic. Glad you took this idea. Cheers!
good information sir
Thank you!
It seems like a lot more than 0.5-1% of people these days are requesting gluten-free dishes. Bread has been a staple food for humans for tens of thousands of years. Do you think a subset of people have always suffered from gluten sensitivity? Is the apparent "popularity" of gluten-free food just over-hype for some people?
Yes, I do think the popularity of gluten-free is over-hype by some people. This article explains it quite well:
Gluten free: Fad diets are all the rage but here's why they shouldn't be. by Ben Locwin | Genetic Literacy Project | February 1, 2017
Gluten in bread is very bad for us, but i think meat is the real killer.
That is a lot of science that is not in my area of expertise. All I can add is that if you look at the average life expectancy over the last 100 years it has only gone up.
Although I look on modern wheat processing techniques with some suspicion it is still hard to ignore the general improvement in the general health of the body public.
Maybe some people's intestines are sensitive to this issue but overall I am not sure whether it is really something for the majority of people to worry about?
Congratulations @davidrhodes124! You have completed some achievement on Steemit and have been rewarded with new badge(s) :
Click on any badge to view your own Board of Honor on SteemitBoard.
For more information about SteemitBoard, click here
If you no longer want to receive notifications, reply to this comment with the word
STOPTo the question in your title, my Magic 8-Ball says:
Hi! I'm a bot, and this answer was posted automatically. Check this post out for more information.