Structure of the Zika Virus Capsid
A member of the Flaviviradae family, the Zika virus is related to other mosquito-borne viruses such as yellow fever, dengue, Japanese encephalitis, and West Nile viruses. It was first isolated in 1947, however, the virus was not really noticed until the 2015-2016 epidemic when it caused numerous cases of microcephaly as well as Guillain–Barré syndrome in South America.
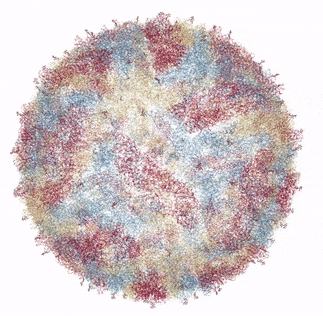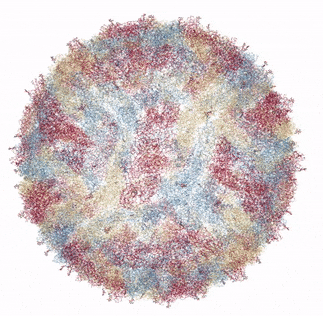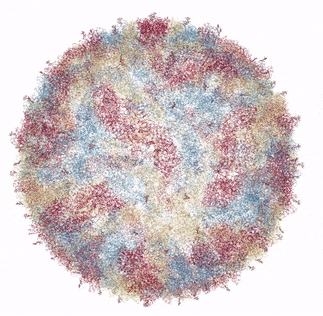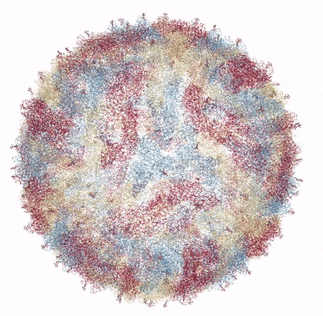
The Zika virus capsid
Source: Own work based on RCSB viewer, RCSB Protein Data Bank
In 2016, a group of researchers from Purdue University collaborating with the National Institutes of Health presented the crystal structure of Zika Virus, determined by Cryo Electron microscopy (Cryo-EM) [1]. Cryo-EM is a method of bombarding a sample with electrons in order to resolve its structure. The mature form of the virus (ie, able to infect humans or insects) was found to be similar in structure to Dengue virus and West Nile Virus. This makes perfect sense as all three are in the flaviviradae family of viruses. In general, the flaviviruses consist of an 11,000 base pair RNA genome (unlike my genome or yours, which is much longer and consists of DNA rather than RNA), as well as various structural and non-structural proteins. The flavivirus capsid is approximately 40 - 60 nanometers in diameter. The Zika virus icosahedral outer shell, or capsid (made up of the capsid protein C) is also surrounded by a membrane studded with a glycoprotein (carbohydrate and protein bonded to each other) E that allows the virus to attach and penetrate a host cell. In addition to the capsid protein C and glycoprotein E, there are numerous other non-structural proteins encapsidated with the Zika virus particle that encode protease, helicase and replicase functions.
Although the Zika virus structure was found to be highly similar to other known flaviviruses, there was a significant difference in the amino acids surrounding the glycosylation site at position 154 of the glycoprotein E, which may account for the differences in transmission and disease between Zika virus and other flaviviruses. This is because the site likely functions as an attachment point of the virus to the host cell.
You, too, can participate in Zika virus research by downloading BOINC and participating in the Open Zika project on World Community Grid. OpenZika is searching for a treatment for Zika virus infection by computationally screening millions of compounds for docking to the Zika virus proteins.. You can even get rewarded for your contributions with Gridcoin!
References
[1] Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, Kuhn RJ. The 3.8 Å resolution cryo-EM structure of Zika virus. Science. 2016 Mar 31. PMID: 27033547.


Can you imagine what we could do if you could bring even 1/10th the energy used to mining BTC to computing for science?
I think Gridcoin is going to explode in 2018, following a great brand update and thanks to the huge, untapped, BOINC user base.
Removal of the team requirement alone I think would propel us to > $1.00 in short order.
Indeed, there would be vast computational power available to scientists. Any problem amenable to computational simulations could be solved very quickly. OpenZika is also only one of several projects on World Community Grid that could make a real difference in people's lives, the other ones being fightAIDS@home, Outsmart Ebola Virus, Computing for Clean Water, Help Stop TB (tuberculosis), Help Fight Childhood Cancer and the Microbiome Immunity Project.
For anyone interested, please consider donating your spare CPU time to science! If you do get involved, reply to this post and I'll help you start collecting Gridcoins for your efforts too!
https://www.worldcommunitygrid.org/