You are viewing a single comment's thread from:
RE: Let's Splice! (My Research, Vol. 3)
Good luck with your research. Are you going to delete the oocyte specific exon using CRISPR?
Cheers!
ian
Good luck with your research. Are you going to delete the oocyte specific exon using CRISPR?
Cheers!
ian
No, we'll try to deactivate the brain specific protein, which means the oocyte exon is already gone.
The gist is that we will add an additional gene, that will only be transcribed together with the brain-specific protein, which leads to a disruption during the translation. It's a bit difficult to explain in a comment, especially without knowing your biological background.
Oh I see now, you want to specifically inactivate the brain specific transcript without disrupting the oocyte transcripts. So like neuronal specific RNAi to knock down the brain specific transcript.
Yes, but we want to completely knock it out, not just down so RNAi is not an option. We'll be putting a different gene in the untranslated region before exon 1, as this region is not there in the oocyte specific version. This will cause the translation to stop after our artificial gene and the brain specific version will never be translated.
I see. But if you put this gene in between the oocyte exon and exon-1 will it affect how the gene splices, i.e. will the oocyte exon splice to exon-1 with this new gene in there? Although the oocyte transcript spices out that intron and doesn't use it, this new gene could interfere with this process and hence you would also be disrupting the oocyte specific protein function.
i.
Splicing will not be influenced actually. Eukaryotic mRNA is monocistronic (I hope I wrote that correctly, English isn't my first language), which means one mRNA can only lead to one protein. After the protein is done (and there was a stop signal), translation stops - no matter how much mRNA ist left! So the protein we introduced is produced and the one we want to knock out is ignored.
@suesa why not flox the gene of interest you want to knockout, and do tissue specific Cre expression by throwing in an inducible Cre driven by a neuron specific promoter? I guess it depends on what model system you're using
That's a good question, I'd have to ask my supervisor. He's been working on this for a while tho and what you suggested is the traditional way, there's probably a reason why we're not doing it like that. It's mice btw.
Is the neuronal specific transcript due to a difference in transcription initiation start site? I though it was due to alternative splicing. Are you knocking in GFP upstream of exon 1 and having a stop codon before the neuronal specific protein?
@suesa I know this is an older post but I still am curious about how you are knocking out the brain specific transcript. I have made a figure of my understanding form your description. But my question is by inserting the Gene X could this not affect the normal splicing of the oocyte specific message?
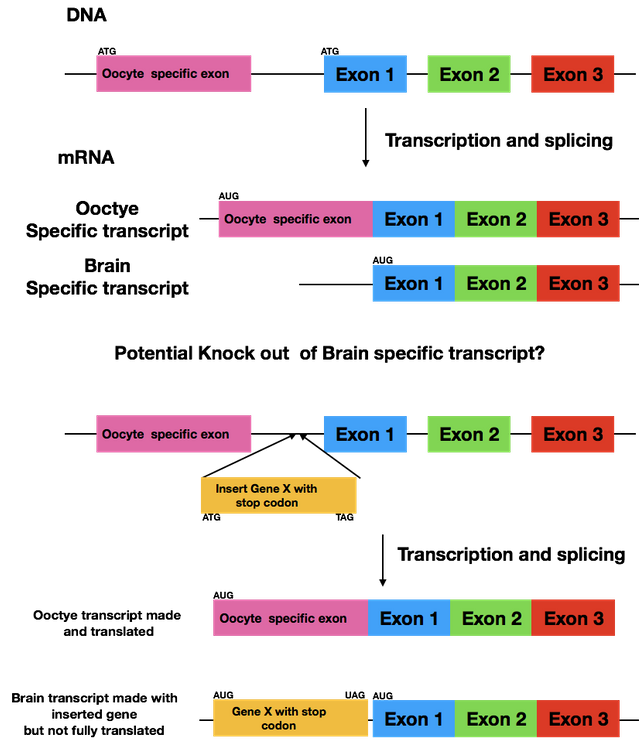
Let's say it like this: we hope it doesn't influence the splicing of the oocyte specific version. The untranslated region we're adding the gene to is only found in the brain specific mRNA. If you're curious how it'll turn out, message me July 3rd as that's one day after I've turned in my thesis.
Ok thanks for the clarification and good luck with your thesis write up and defence!
Here is a practice defence question for you:
Why don't you just delete the intron between the oocyte specific exon and exon-1 ( to make a combined larger exon) to "knock out" the brain specific transcript?
btw what is the proper protocol or etiquette for messaging users? I just discovered Discordapp.com and should I contact users there or in the comments section of their posts?
Cheers!
ian