The personal and clinical utility of polygenic risk scores – a summary
In my lab, every Monday at noon, we have a meeting called Lunch and Learn that consists of anything from informal discussion of an interesting topic to postdoc interview talk. Last month, my PI suggested reserving a L&L to discuss a review paper on polygenic risk scores (PRS). As a newbie, I have not had a chance to present at L&L so I volunteered to lead the discussion. Full disclosure, I have not worked with GWAS data before, let alone computing PRS, but I thought this would be a good opportunity for me to learn more about the topic.
The personal and clinical utility of polygenic risk scores reviews the evidence supporting polygenic risk profiling is helpful in identifying high risk individuals of Alzheimer disease, breast cancer, prostate cancer, coronary artery disease and type 2 diabetes mellitus.
Motivation
Despite having been criticized of having low utility due to very small phenotypic variance explained at the population level (or little power in discriminating diseased vs. non-diseased individuals), Genome-wide Association Studies (GWAS) have recently showed potential to identify a subset of individuals at high risk of disease (genetic + clinical risk factors).
Heritability in a population vs. individual disease risk
A breakdown of disease risk includes environmental exposures, lifestyle factors and genetic susceptibility, which can be quantified by heritability of that disease in the population. However, the authors provided a nice example to demonstrate the difference between heritability in a population and individual disease risk:
Although the total heritability explained by BRCA1 and BRCA2 variants is low [due to low prevalence of mutations], BRCA1 and BRCA2 testing can identify a subset of individuals whose absolute risk of disease is significantly higher than that of the average individual in the general population.
In brief, even though at the population level, analyses in GWAS may not be able to discriminate the cases from the controls (area under the receiver operating characteristic curve close to 0.5) but provide reasonable disease risk stratification of the population, which is helpful information from the individual perspective.
Common variants vs. rare variants
Pedestal of this review is the concept of polygenic, a genetic architecture where common low-risk loci contribute to the occurrence of disease in an individual. The paper stated that statistical modeling and empirical results corroborate the makeup of genetic architecture for many common adult-onset diseases:
- familial form
- 1–10% of disease incidence
- highly penetrant rare variants
- small set of genes
- nonfamilial form
- common variants of small effects, throughout genome
- smaller contribution from rare variants of moderate effect, in genes driving familial disease
In other words,
The genetic architecture of common adult-onset diseases is likely a continuum of common low-risk to rare high-risk genetic variants that can act cumulatively to drive overall risk in any single individual.
However, I did feel like the authors downplayed the role of rare variants. They referred to a recent large-scale, comprehensive GWAS of CAD and another on type 2 diabetes mellitus that suggested the majority of disease heritability is likely explained by common variants with small effect sizes. In either study, little evidence was found for low-frequency risk variants with moderate/intermediate effects despite being well powered to detect such associations. This common disease, common variant hypothesis will probably be a big discussion point.
To look up: copy number variants.
What is polygenic risk score?
Ultimately, polygenic risk score (PRS) is commonly calculated as the weighted sum of the number of risk alleles carried by an individual based on the loci and measured effects, as reported in the literature.
To read: an in-depth review on considerations for the development of PRSs
What can we do with polygenic risk scores?
Three things: intervention, screening, and life planning.
Therapeutic intervention
Individualized management of disease is central to the philosophy of precision medicine, with genetic factors often invoked for this strategy to personalize health care.
Example:
- CAD PRS helps ID individuals who receive greater benefit (e.g., heart attack risk reduction) from statin therapy initiation.
- CAD PRS reclassified ~12% intermediate risk individuals to high risk.
Disease screening
PRS helps correct the age at which an individual should start screening for breast cancer to balance the average risk of breast cancer and the risk of harms due to false positive mammography results.
On the other hand, prostate cancer PRS could help prioritize screening subgroup at high risk.
Life planning
How does PRS-informed life planning different than conventional life planning? In other words, don't we all already know what a healthy lifestyle looks like? Regardless of genetics, don't we all know that smoking is bad for you? Yes, but...
Theoretically, clarifying a high-risk individual's perception of their susceptibility to disease and quantifying the benefits of healthy behaviours would be one among many effective tools for inducing and maintaining behaviour change. [The Handbook of Health Behavior Change]
This has been shown to be effective in breast cancer and coronary artery disease cases. Studies have also shown that the Alzheimer PRS could dramatically stratify individuals by average age of disease onset.
Caveats
Imperfect correlation with causal genetic factor(s) → Uncertainty in variant’s estimated effect size → Poor transferability → Inequities
Future directions
- Capture uncertainty of individual's risk estimates → distribution
- Integrate with familial risk
- Whole-genome prediction
- Dynamic model: demographics, lifestyle, clinical risk factors
- PRS models via ML: data and interpretation
In conclusion
Even though the abstract started out with a defeatist tone stating GWAS data provides little useful information in predicting disease status, the entire review showed promises in utilizing PRS to inform individuals of their genetic disease risk, which may lead to proper modification of lifestyle, timely screening and intervention.
Oversights
- “As for” should be “Like” in the following sentence:
As forLike type 2 diabetes mellitus, rare variants of intermediate effect size in genes associated with familial breast cancer are known to have an important role in breast cancer predisposition.
- Torkamani et al's review paper showed the impact of the reduction of modifiable risks in total risks by referring to a breast cancer study. They paraphrased one of the findings:
For breast cancer, if healthy lifestyle choices were preferentially targeted to and employed by women in the top decile of genetic risk, an estimated ~20% of all preventable breast cancer cases would be avoided.
This sentence is rather confusing, since I'm not sure whether the 20% refers to number of cases across all deciles or just from the top decile. When looking into the referred paper for findings, I found this confusing-as-heck table:
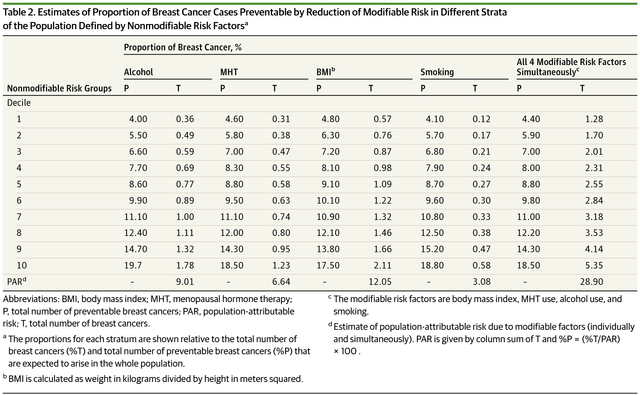
Essentially, groups are defined based on non-modifiable risk and the authors of this study estimate how much overall risk can be reduced in these groups by changing the modifiable factors, like quitting smoking or quitting snickers.
This is what the original study has to say about this table:
Overall, we estimated that up to 28.9% of all breast cancers could be prevented if all white women in the US population were at the lowest risk from these 4 modifiable risk factors. Nearly one-fifth of these total preventable cases arise from the subpopulation in the top decile of nonmodifiable risk. In contrast, only about 4% of the preventable cases arise from the population in the lowest decile of nonmodifiable risk.
If we look at the last column for the 10th decile, only ~5% cases would be avoided when healthy lifestyle choices were employed by top decile individuals. I think Torkamani et al might have misinterpreted the result here. Please let me know if you have a different opinion!
Thoughts
It seems to me that many people are computing PRS due to its simplicity and tremendous utility from the individual standpoint. There are of course still debates on PRS's statistical validity and clinical practicality. One co-author of this study, Eric Topol, recently tweeted:
The polygenic risk scores, like the wealth of key pharmacogenomic findings, sits in the corpus of medical research orbit that have yet to benefit the public. It's time to change that.
Unfortunately, I feel like it will be a while until most of these findings get out of the research orbit.
If you’re interested, please feel free to take a look at my slides for the discussion (work-in-progress).
If we have time at the L&L, we will also discuss another specific study on utilizing PRS that has got some press lately:
I hope to write another summary for this paper soon.
Until then!
One word only ... respect!
haha @zorank thank you, but I didn't write these papers. I was merely trying to gain some knowledge on the topic.
Ok, I get it, but still. One needs to have some knowledge to read and understand papers like that.