Circulating Tumor DNA: A non-invasive way to detect cancer early
How does tumor DNA get in the bloodstream?
All humans (and most creatures with a circulatory system) have DNA floating around their blood. When cells die within or nearby blood vessels, they often release their DNA into the circulatory system and it can persist for some time before nucleases degrade it. This type of DNA is known as cell free DNA (cfDNA) and is usually present at low and stable levels.
When a cancer grows inside the body, the levels of free DNA in the blood can dramatically increase (1). This occurs for two main reasons. For one, tumors usually grow adjacent to blood vessels because they need the nutrients from blood to maintain a high rate of growth. The rapidly growing tumor cells die quickly as well, either through apoptosis or because the fast growth stresses them to the point of death. When these cells die, they release their DNA and much of it migrates into blood vessels. It’s called circulating tumor DNA because it is essentially DNA from tumor cells that circulate through the body.
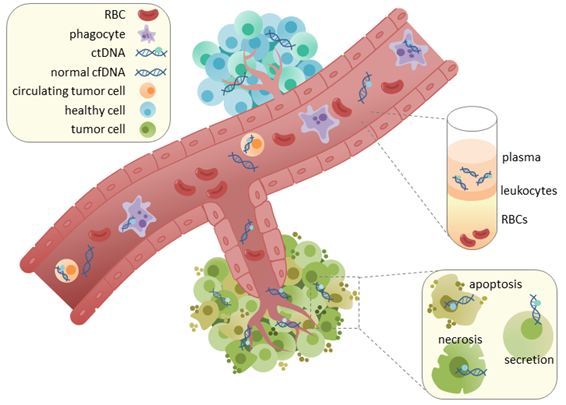
Diagram explaining how ctDNA gets in the blood. DNA is released from cells by apoptosis, necrosis, or secretion and migrates to blood vessels. From (2)
Why do we care about this DNA?
Scientists have theorized about the benefits of circulating tumor DNA for a long time. The initially discovery of ctDNA was in 1977 (3) when scientists showed that cell free DNA levels were elevated in cancer patients, and these levels decreased after radiation therapy. Even back then it was obvious that simply analyzing blood samples for elevated free DNA levels could be used to detect cancer. It wasn’t a perfect method as cancer won’t always release DNA into the blood, but the fact that all that was needed was a simple blood sample was critical. Drawing blood is a non-invasive technique. It doesn’t pose much risk and, most importantly, it won’t physically disrupt a tumor which could cause it to spread further (2). In addition, multiple blood samples can be taken over time to track a cancer’s progression or regression.
Circulating tumor DNA has rapidly grown in importance over the past 10 years as our DNA detection methods have improved. Instead of just looking at total DNA levels, scientists could begin to read the sequences of the DNA released into the blood (4). They could begin to detect cancer biomarkers, a catch-all term that can be used to refer to DNA sequences indicative of cancer.
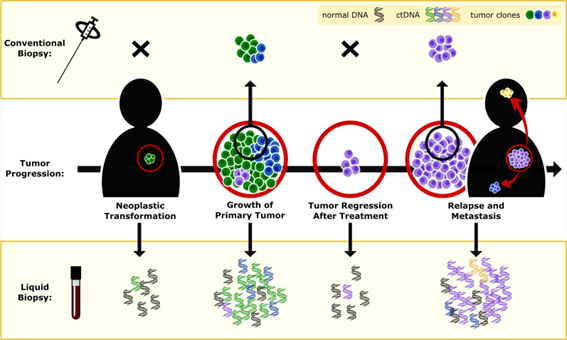
Liquid biopsies (aka blood samples) can be analyzed to detect a tumor’s ctDNA. Analysis of a patient after treatment can show whether the tumor comes back and also help identify any new mutations the tumor has accrued (2).
Not only does this improve the accuracy of using ctDNA to detect cancer, but it can help doctors determine the exact type of cancer affecting the patient. As our knowledge of cancer biomarkers becomes more robust and our next generation sequencing methods improve, we can more accurately identify the cancer affecting a patient and tailor treatment specific to that cancer. This is often referred to as precision medicine and is one of the most sought after goals of modern medical research.
What have we accomplished so far?
OK now for the exciting part where I do a deep dive into a current paper. Well… this is exciting for me at least. If you aren’t titillated by graphs and charts, feel free to skim.
Most of the exciting applications for ctDNA have yet to be put into practice. Some clinical tests do focus on detecting this DNA and a few tests that will begin to leverage next generation sequencing to detect cancer in ctDNA are expected to be available soon (5). However, the major advancements in this field are still done in laboratories rather than hospital rooms. These studies are often longitudinal as well, since researcher’s need to chart the development of a cancer alongside the changes in ctDNA over time. Although this means progress is slow, most research is done directly on human patients since the blood analysis is non-invasive.
A great example of this kind of research is described in the Nature paper “Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy” (6). In this study, researchers focused on a group of patients treated for Triple-negative breast cancer. This type of breast cancer is uncommon, but particularly malignant and has a high likelihood of relapse. The researches wanted to determine if they could analyze circulating tumor DNA to detect a relapse before conventional methods were able to do so.
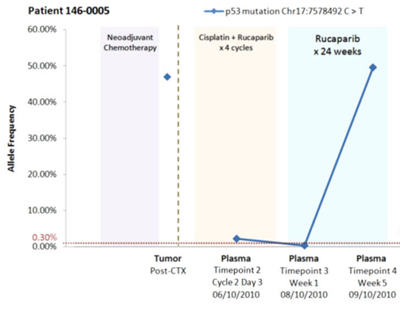
Relative increase of a cancerous mutation (allele) in the cell free DNA of a patient’s blood. The significant increase after 24 weeks predicted a resurgence of the cancer (6).
To do this, the researchers took tumor samples from 38 patients that had received chemotherapy, surgery, and radiotherapy designed to eliminate the disease. They were able to detect cancer-specific mutations in 33 of these tumor samples. They then looked at blood samples and attempted to find circulating tumor DNA matching the mutations in the tumor. They found a match in 4 of the 33 patient samples. What’s interesting is that all four of these patients had rapid recurrence of the cancer and died within a year. In the patients without any detected circulating tumor DNA, the rate of recurrence was 28%. In other words, the presence of ctDNA guaranteed cancer resurgence, but even patients without detected ctDNA could experience a relapse.
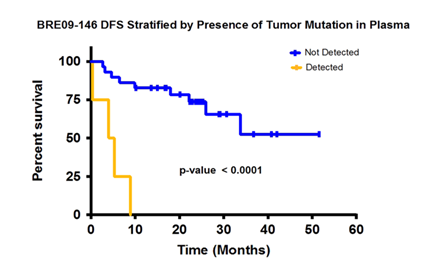
All the patients with detected ctDNA died within 10 months. The rest of the patients had a much better survival rate (6).
It’s a bit depressing that this major nature paper only showed that it might be able to predict a patient’s death ahead of time, but its implications are huge. Having another tool that can help predict the success or failure of a cancer treatment more rapidly can dramatically increase survival rates and can hasten treatment. It’s also important to note that the current limitations of genome sequencing technology and cancer bioinformatics were two of the biggest factors holding back studies like these. These two factors are constantly improving which should make analysis of ctDNA much easier in the near future.
Studies similar to the one described above are ubiquitous the field at the moment. Paper titles like “Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer” (7), and “Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers” (8) are common. Although these studies have yet to transfer to routine clinical application (9), the success of them suggests that We may expect analysis of ctDNA to be one of the first widely available tools for personalized medicine.
Images
All images were taken either directly from the publication referenced, from Wikipedia Commons, or by myself. If any image owner has an issue with this article, please contact me and I will address the issue.
References
(1) https://www.sciencedirect.com/science/article/pii/S0092867417301150
(2) https://en.wikipedia.org/wiki/Circulating_tumor_DNA
(3) https://www.scopus.com/record/display.uri?eid=2-s2.0-0017360626&origin=inward&txGid=f75a328d97809828b73f844d1d546f5f
(4) http://cancerdiscovery.aacrjournals.org/content/6/5/479
(5) https://www.sciencedirect.com/science/article/pii/S1672022917300487
(6) https://www.nature.com/articles/s41523-017-0028-4
(7) http://stm.sciencemag.org/content/7/302/302ra133.short
(8) http://www.gynecologiconcology-online.net/article/S0090-8258(16)30170-6/abstract
(9) http://cancerdiscovery.aacrjournals.org/content/6/5/479.short
SteemSTEM
I’m a proud member of the @steemstem community and I encourage you to join. SteemSTEM is a community driven project which seeks to promote well written/informative Science Technology Engineering and Mathematics postings on Steemit. The project not only curates STEM posts on the platform through both voting and resteeming, but also re-distributes curation rewards as STEEM Power, to members of Steemit's growing scientific/tech community.

When I read this I was like "smart!" They also do angiogenesis right?
If a cancer doesn't do that, that means it's local? Or are there other ways for it to metastasize? I think via lymph nodes?
Overall a great idea for early detection. Or at least that's how I imagined it at first: blood tests getting done on patients who haven't been diagnosed with cancer. But it seems this is used on patients who have been treated for cancer, in order to detect relapse. Hopefully in the future it'll just be part of routine blood exams?
I think its too early for this to be a routine blood test, but in the future we might see it become the norm. It a tumor doesn't release DNA that might mean its local or benign, but cancer is so varied it could mean a lot of things. I think time and better sequencing tech will shed a lot of light on this area.
Great article, I'm not medically minded but you made it easy to understand. For a layman, the study detecting the reoccurring cancer with a ~60% success rate is pretty impressive. And maybe I'm wrong here but it sounds like blood based testing could bring better detection to people with limited access to hospitals. Exciting stuff!
That's very true. Thanks for reading and thanks for the compliment :)
@originalworks
Great post and thank you for contributing to your community and here on Steemit. I love to read scientific articles, especially from people, not some pages...
I'm sad that more and more people have cancer, but I'm thankful for people like you who are a great helpers to others...
Enjoy your place in a new city ;) Luka
Thanks! The SteemSTEM community is great at encouraging us to write scientific articles and its always nice to see people reading them.
Yeah, I started to write some scientific articles but I don't have that much time now even to read anything that's not on Steemit.. Kinda sad about that, but this article and other stem articles are saving my day! :D
Keep coming bro!
I remember that someone had written something along these lines (cancer detection through DNA analysis) on steemit. Was it you? But it is the first time I am reading about the potential applications of it. I hope this will make the future brighter!
I usually search steemit for any similar articles before I choose a topic. In this case I found a few, https://steemit.com/science/@altherion/paper-discussion-3-a-prevention-for-cancer and https://steemit.com/science/@bankopm/cancer-can-be-predicted-years-ago, but neither go into the technique or science in much detail so I figured I'd be covering enough new ground. At any rate, this is my first time covering the topic, and I hope it makes the future brighter as well!
Please do not misunderstand my comment. It was not a criticism. The only thing I wanted to point out was that reading your post triggered some connection in my mind. Yours is of course the deepest of all on this topic, and thanks for putting this up ^^
PS: altherion's was the one I read.
No offense taken. I often get the same connection or sense of deja vu reading some articles.
Yea everything is true... It also help doctors determine the optimal cancer treatment for a patient.