CHEMISTRY OF NUCLEIC ACIDS: DNA REPLICATION AND DNA REPAIR #4
By Nastypatty - CC BY -S.A 4.0, Link
Hi again and welcome to another great series of CHEMISTRY OF NUCLEIC ACIDS: DNA REPLICATION AND DNA REPAIR, on our last post we said RNA is a polymer of ribonucleotides of Adenine, Uracil, Guanine and Guanine and Cytosine, joined together by 3’ 0 5’ phosphodiester bonds. we also looked at the structures, types and characteristics of RNA. Now we will be going into another post, and if you miss the previous ones here is a link to:
NUCLEIC ACIDS: DNA REPLICATION AND DNA REPAIR #1
CHEMISTRY OF NUCLEIC ACIDS: DNA REPLICATION AND DNA REPAIR #2
CHEMISTRY OF NUCLEIC ACIDS: DNA REPLICATION AND DNA REPAIR #3
By Vossman - CC BY -S.A 3.0, Link
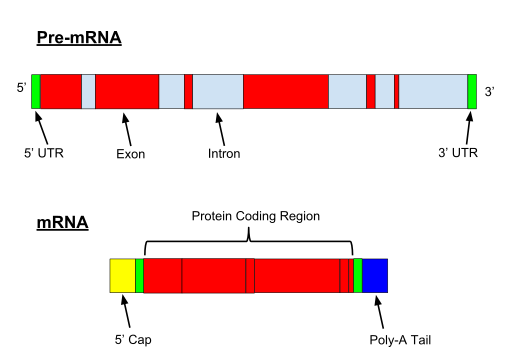
What is PRE-mRNA?
PRE-mRNA
- Pre-mRNA is converted to m-RNA
- Pre-mRNA has regions called as ‘’introns’’ transcripts the sequences not required (inactive) and ‘’exons’’ transcripts (active portion required for translation.)
- Approximately 80% of length of pre – mRNA is removed as ‘’introns transcripts’’ and only 20% of Pre-mRNA, the ‘’exons transcripts’’ are spliced to form the m-RNA.
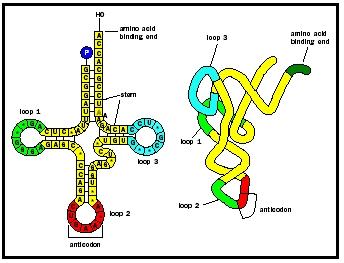
TRANSFER RNA OR T-RNA
- Primary structure of t-RNA t-RNA molecules consist of approximately 75 nucleotides. Their bases include adenine, guanine, cytosine, uracil, pseudouridine (ѱ) or uracil 5-ribofuranoside and thymine are present in one loop.
- Secondary structure of t-RNA Each single stranded t-RNA molecule remains folded to form a clover-leaf secondary structure. These folds of the secondary structure are stabilized by H-bonds between complementary bases in different portions of the same strand, these double stranded hetical structures are called as stems.
All t-RNA molecules contain 4 main arms or loops. Which are:
Acceptor arm
This is consists of unpaired sequences of cytosine-cytosine-adenine at the 3’ end also called as acceptor end. The 3’ –OH terminal of adenine may bind with the cl-COOH of a specific amino acid and carry the latter as an ‘’amino acyl-t-RNA complex’’ to ribosomes for protein synthesis. The acceptor arm is borne by a base-paired acceptor stem whose bases are hydrogen bonded with the last few bases at the 5’ end of t-RNA.
Anticodon arm This is another on paired and non-bonded loop carrying specific sequences of three bases constituting the anticodon. The bases of anticodon are hydrogen bonded with three complementary bases of codon of m-RNA. The base pair stem leading to anticodon loop is called the ‘anticodon stem’.
D arm
The third is the D-arm because it contains the base dihydrouridine.
T ѱ C arm
Contains Thymine, Pseudouridine and Cytosine.
Variable arm or extra arm
Extra arm is most variable structure of t-RNA and it forms the basis of its classification.
T-RNA can be classified into:
- Class I t-RNA About 45% of all t-RNA belong to this class and have 3-5 base pairs in its extra arm e.g., Ale-t-RNA.
- Class 2 t-RNA This forms about 25% of total t-RNA and has 13-21 base pairs in a long chain e.g., Phe-t-RNA. They also have a stem loop structure.
The secondary structure of t-RNA is mainly maintained ny the base pairing in these arms and this is a consistent feature. The number of base pairs in these arms remains fixed or varies within a specified range as follows:
L-shaped tertiary structure of t-RNA
The L-shaped tertiary structure is formed by fertile
PRIMARY AND SECONDARY STRUCTURE OF t-RNA Folding of the cloverleaf due to hydrogen bonds between T and D arms. The base paired double helical stems get arranged into two double helical columns, continuous with and perpendicular to one another. The tertiary structure locates the amino acid acceptor end and the anticodon at the farthest ends of the two columns.
Ribosomal or r-RNA A ribosome is present in the cytoplasm and is a nucleoprotein. It is on the ribosome that the m-RNA and r-RNA interact during the process of protein biosynthesis. Ribosomes contain the third type of RNA known as r-RNA. The r-RNA forms 80% of the total cellular RNA.
- Ribosomes possess a sedimentation coefficient of 80s with a mol. Wt. 4.2 ×106. Mammalian ribosomes are made up of two subunits, larger one with 60s and mol. Wt 2.8 × 106 and smaller subunit with 40s and wt of 1.4 × 106. The 60s subunit carries 60% of r-RNA and is a combination of 5s r-RNA, 5.8s r-RNA 28s r-RNA. The 40s subunit carries the r-RNA and 33 different proteins All the r-RNA molecules except 5s r-RNA are processed from a precursor of 45 s r-RNA.
- Mammalian 5s, 5.8s, and 28s r-RNA are made up of about 120, 160, 1900 and 4700 bases respectively. The bases consist and uracil and a few pseudouridines.
- The function of r-RNA in ribosome particle is still not clearly understood. However, they are necessary for ribosomal assembly and seem to play key roles in the binding of m-RNA to ribosomes and its translation.
Regulation of synthesis of Ribosomes (r-RNA) Living cells exert so-called ‘’stringent control’’ over syunthesis of ribosomes. Ribosmes are not produced unless they are going to be used by the living cells. In the non-avaialbilty of amino macids cells stop the synthesis of ribosomes and when aminoacids are available the control is relaxed and synthesis takes place.
Mechanism
Two nucleotides exert the control. They are:
- ppGpp: Guanosine diphosphate 2’ *or 3’) diphosphate
- pppGpp: Triphosphate derivative.
When amino acids are not available, these nucleotides can turn off the synthesis of r-RNA (ribosomes). These nucleotides can be produced enzymatically by ribosomes in prescence of ATP. GDP, m-RNA and t-RNA. Evidence
These two nucleotides have been shown to be present as two spots in electrophoretogram, when cell extracts of normal cell under ‘stringent control’ is subjected to paper electrophoresis. These two spots have been named as ‘’magic spot1’’ and ‘’magic spot 11’’ respectively.
Miscellaneous RNAs Sn RNA:
Small nuclear RNA SnRNA are large number of small stable RNA species found in eukaryotic cells. They range in size from 90-300 nucleotides and are present in `100,000-1000,000 copies per cell. Small nuclear ribonucleoprotein particles Sn RNP called as Snurps are involved in gene regulation. There are U1, U2, U3, U4, U5, U6 and U7 types of snurps with different lengths of nucleotides. Some of them have been found to perofrm specific funtions.
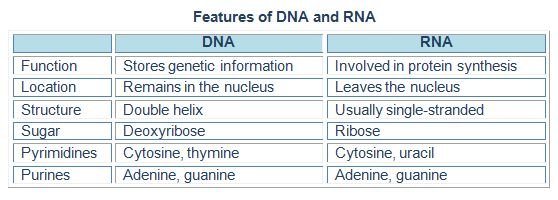
Differentiating features of DNA and RNA is given in a tabular form
Topic will be continued.
Thank you all for reading till next time.
References
- Nature.com: Transfer RNA / tRNA
- Quora.com: How many types of tRNA are there?
- Ncbi.nlm.nih.gov: The Three Roles of RNA in Protein Synthesis
- Sciencing.com: Why Are There Many Different Types of tRNA Molecules?
- Thoughtco.com: What Are the Three Types of RNA? What Are Their Functions?
- Khanacademy.org: tRNAs and ribosomes
- Britannica.com: Transfer RNA
- Biology.stackexchange.com: Why is tRNA called an adaptor molecule?
- Upendrats