Salads Under Siege, Episode 6: Populations Racing
Here I will discuss the evolutionary forces that have sustained the plant-pathogen running game for thousands and thousands of years.
In my previous posts, I explained how both plants and pathogens have acquired and evolved elaborate weapons arsenal to fight each other. The rewards? Growth, reproduction, and transmission of the best genes to the next generation. Most people would think that such rewards are obvious and perhaps not particularly imaginative. But this struggle for survival and the transmission of the best possible genes to offspring is what explains so many of the observations made in biology.

Figure 1. A population racing. Pathogens and plants alike are always racing each other, vying to be the fittest. Source
As I mentioned before (and I will, again and again), passing on a gene to the next generation is critical. The question therefore is, what determines whether a gene is going to be passed on? A gene can survive and prosper if it gives a particular advantage to an organism in nature. Genes can mutate when they are replicated and when this happens, new variants arise. These variations can cause changes in amino acid sequence, and when that occurs, protein functions are altered. In many cases, these alterations are minor or non-consequential. In other cases, the protein seizes to function, and when this negatively impacts the organism, it will become less competitive, and the gene is likely to disappear from a population.
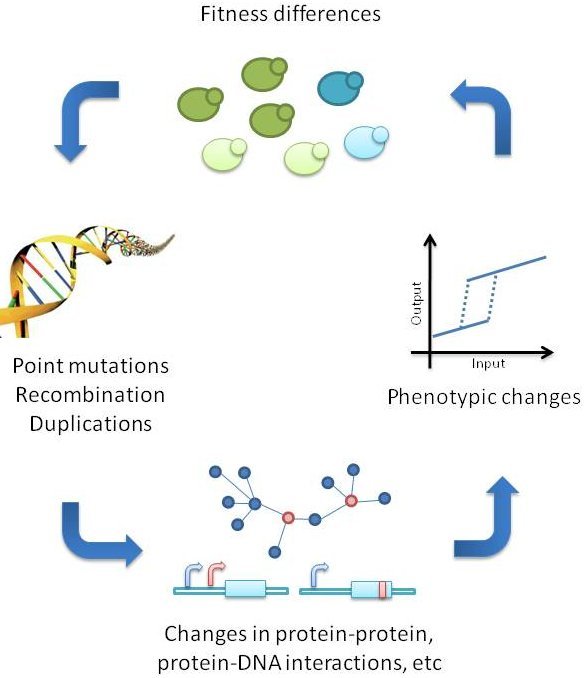
Figure 2. Illustration showing how mutations in genes can lead to changes in protein sequence, function and ability to interact with other proteins/compounds. Mutations are often random, but tend to be selected for if there is a consequence to the fitness of the organism carrying the mutated gene variant Source
Let's go back to plant-microbe interactions. Say, there is a pathogen that can infect all members of a diverse plant population. The pathogen will infect with high frequency, kill most plants and thereby, eliminate many genes from the gene pool that are of no use to the plant in terms of defense against the pathogen. However, the plant population is not standing still. Individuals that carry mutations in one or more genes may arise. A gene version will be unique initially per definition as it arises in an individual and depending on its contribution towards plant fitness may disappear again. However, if the mutation has a positive effect on the plant's ability to withstand the pathogen that is rampaging the population within a species, it will give it a distinct advantage. The resistant plant will produce offspring, many of which will have the new improved gene variant. This offspring will be more successful again initially, and consequently, the frequency of this gene variant will increase. In other words, plants with the gene will survive and selected for, and therefore, the gene variant is under positive selection. Indeed, population studies and functional analyses have identified a large repertoire of receptor coding genes that can detect effector molecules that pathogens produce and need to infect plants. In other words, random gene evolution events have generated an arsenal of proteins able to counter pathogen effectors (and thus disable infection) by initiating strong immune responses. Innovation allows plants to keep up with pathogens. Critically, however, we can only attribute this feat to plant populations.

Figure 3. Illustration of the process that drive host-pathogen co-evolution. Both pathogen and host need to be successful and in try do so, will mediate the selection of genotypes that are either resistant (plant) or become virulent on a previously resistant plant (pathogen). Source
So what does this mean to the pathogen? From a population point of view, the recognition of an effector by a plant receptor has little consequence initially. Most plants will be susceptible, and most infection events will, therefore, be productive. However, when the proportion of resistant plants increase, chances of survival will decrease. Pathogen individuals that carry a recognizable effector get eliminated from the population. In other words, negative selection is taking place against the effector, where the new receptor coding gene applies the pressure and confers resistance. What could the possible consequences be?
Well, as you can imagine. The pathogen population will have to find a way to circumvent recognition, and natural variation here is key. As the pathogens are not genetically identical, some individuals could already lack the recognized effector gene but struggled to compete with more virulent strains that have the effector. The presence and high frequency of the receptor in the plant population suddenly hand an enormous advantage to those strains that lack the effector, allowing them to prosper and multiply as the effector+ strains are decimated. Amongst those individuals, there will be variation, and this allows selection of the fittest individuals yet again. When disease pressure builds despite the presence of the receptor in the plant population, selection will retake place, favoring those plant genotypes that have receptor variants able to recognize other effectors. This boom and bust cycle in both host and pathogen ensures gene variation, functional innovation and in some cases, speciation, providing the beautiful variety we see today.
Why is this important?
Unfortunately, it also means that breeding elite crops (deliberately combining good genes into a single crop plant genotype) is an exercise that can yield resistant plants, but that such plants only offer protection for a limited amount of time. Pathogen strains will emerge that can defeat elite resistant cultivars. Not surprisingly, the literature is littered with examples that demonstrate the pathogens ability to evolve and overcome immunity (Armstrong et al., 2015). Such studies have helped unveil a variety of mechanisms that pathogens can employ to rid of an effector molecule. These include gene mutation, deletion (Na et al., 2013) and even silencing (loss of gene expression) (Qutob et al., 2013).
In my next post, I will describe one innovation that plants have evolved to fight off pathogens.
REFERENCES:
Armstrong, M. R., Whisson, S. C., Pritchard, L., Bos, J. I., Venter, E., Avrova, A. O., ... & Hamlin, N. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proceedings of the National Academy of Sciences of the United States of America, 102(21), 7766-7771.
Na, R., Yu, D., Qutob, D., Zhao, J., & Gijzen, M. (2013). Deletion of the Phytophthora sojae avirulence gene Avr1d causes gain of virulence on Rps 1d. Molecular Plant-Microbe Interactions, 26(8), 969-976.
Qutob, D., Chapman, B. P., & Gijzen, M. (2013). Transgenerational gene silencing causes gain of virulence in a plant pathogen. Nature communications, 4, 1349.
I hope you will find these pieces interesting. Your vote and follow help me open up the exciting world of plant biology and pathology to you.
Thank you @foundation for this fantastic SteemSTEM gif

This is raw science with nice sources as back ups. Nice picture illustrations and good explanation. Good job done on this post. Well done!
Thank you for your comments. I hope there are more posts to follow!