Salads Under Siege, E8: Pathogen Decoys and traps
In my previous post, I explained how plants could innovate and build traps to fend off pathogens. I described how plants produce molecular mimicks to trick pathogen effectors to the wrong target (decoy). Evolution of genes in the plant genome has led to fascinating innovations that thwart pathogens and help fight off infection.
It is a mistake, however, to think that pathogens will sit by idly. Indeed, pathogens have evolved their own set of molecular props in a bid to remain able to colonize plants. Here, I will describe one of those pathogen accessories, akin to decoys found in plants. Its a protein called PsXLP1 (for Phytophthora sojae XEG1-Like Protein).
It all started when researchers studying effectors from the pathogen Phytophthora sojae identified a secreted Xyloglucan-specific EndoGlucanase (PsXEG1) and demonstrated that this protein is an effector, required for infection by degrading cellular structures (Yoshizawa et al., 2012, Ma et al., 2015). While this protein acts in favor of the pathogen by degrading host barriers, PsXEG1 also is a Pathogen Associated Molecular Pattern (PAMP), triggering immune responses in its host Soybean (Glycine max) (Ma et al., 2015). It is clear therefore that this protein must greatly help the pathogen (why else would a plant target it?).
These results combined with the fact that plants encode secreted Glucanase inhibitors led some researcher to try and identify PsXEG1 inhibitors in the host. Immuno-precipitation experiments led to the identification of soybean Glucanase Inhibitor Protein 1 (GmGIP1) (Ma et al., 2017). When over-expressed, GmGIP1 limits P. sojae infection, and biochemical studies showed it to be a bona fide inhibitor of PsXEG1. Thus, soybean expresses an inhibitor that can counteract the activity of a secreted pathogen glucanase. These observations raise an important question though:
IF GmGIP1 can inhibit PsXEG1 and limit infection when present at higher levels, then how can P. sojae still happily infect soybean plants?
This is where another protein, related to PsXEG1, comes in. Genome sequencing has allowed comprehensive views of gene repertoires in pathogens. Searches against the predicted P. sojae proteome therefore allowed the identification of PsXEG1 homologs, some of which are expressed during infection (Ma et al., 2017). These homologs were cloned, expressed and tested for their ability to bind GmGIP1. Low and behold, one protein was identified (PsXLP1) that was able to bind GmGIP with high affinity but did not have any glucanase activity. PsXLP1 happens to sit adjecent to PsXEG1 in the P. sojae genome and is expressed in the same way. These results suggested that perhaps PsXLP1 was duplicated and evolved the ability to capture the host protein (GmGIP) before it could act on the active pathogen glucanase (PsXEG1) and allow the glucanase to function.
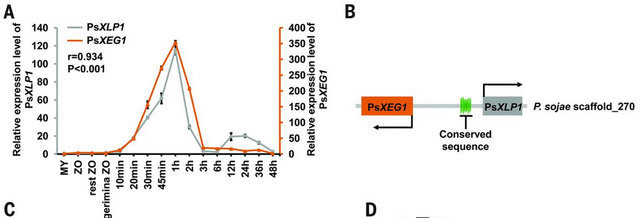
Figure 1. Expression patterns and location of PsXlp1 and PsXeg1 in P. sojae (Modified from: Ma et al., 2017, Figure 2)
Competition and functional assays turned out to confirm this hypothesis. PsXLP1 was able to displace GmGIP from PsXEG1 in competition assays. Loss of PsXLP1 also mimicked the GmGIP over-expression phenotype, loss of P. sojae virulence. From this work, a model now has been proposed in which pathogens have adopted the decoy approach in a bid to fool the host defensive apparatus and protect its most important assets (enzymes that can break down host barriers).
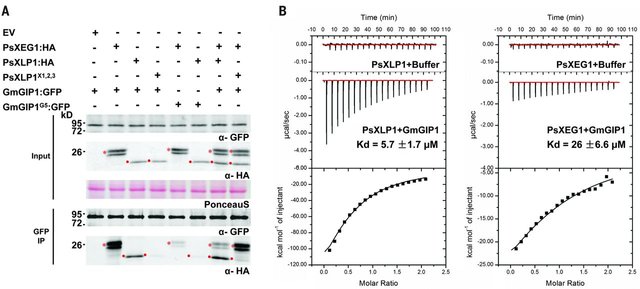
Figure 2. Competition assays demonstrate the PsXLP1 can out-compete the host glucanase inhibitor GmGIP1 to protect the essential pathogen encoded glucanase PsXEG1. Source: Ma et al., 2017, Figure 3
Why is this important and interesting?
Well, there is little question that host and pathogens are locked into an arms race. While it was previously established that plants can evolve and deploy decoys to fight pathogens, it is now clear that pathogens can do this as well. The implication is that when it comes to host-microbe interactions, there are now good reasons to assume that if a host has evolved a particular strategy, that pathogens will follow suit and vice versa. This means that studying host biology can reveal new concept that apply to pathogens and the other way around!
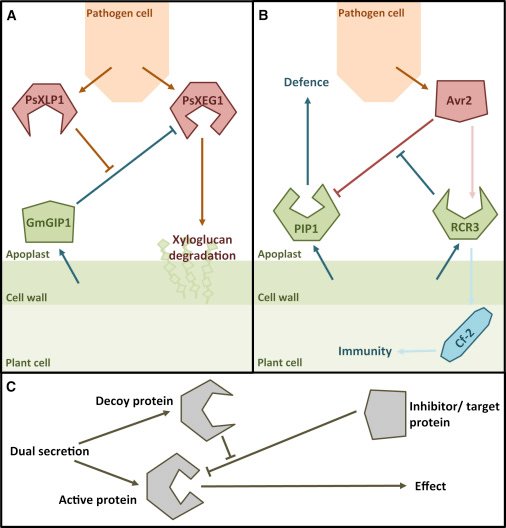
Figure 3. Conceptual model explaining analagous decoy strategies in plants and pathogens. From: Cummins and Huitema (2017)
REFERENCES
Cummins, M., & Huitema, E. (2017). Effector–Decoy Pairs: Another Countermeasure Emerging during Host–Microbe Co-evolutionary Arms Races?. Molecular Plant, 10(5), 662-664.
Ma, Z., Song, T., Zhu, L., Ye, W., Wang, Y., Shao, Y., Dong, S., Zhang, Z., Dou, D., Zheng, X., Tyler, B.M., Wang, Y. (2015). A Phytophthora sojae Glycoside Hydrolase 12 Protein Is a Major Virulence Factor during Soybean Infection and Is Recognized as a PAMP. Plant Cell. Jul;27(7):2057–2072. doi: 10.1105/tpc.15.00390
Ma, Z., Zhu, L., Song, T., Wang, Y., Zhang, Q., Xia, Y., Qiu, M., Lin, Y., Li, H., Kong, L., Fang, Y., Ye, W., Wang, Y., Dong, S., Zheng, X., Tyler, B.M., Wang, Y. (2017). A Paralogous Decoy Protects Phytophthora Sojae Apoplastic Effector PsXEG1 From a Host Inhibitor. Science. Jan 12;355 (6326),710-714. doi: 10.1126/science.aai7919.
Yoshizawa, T., Shimizu, T., Hirano, H., Sato, M., Hashimoto H. (2012). Structural basis for inhibition of xyloglucan-specific endo-β-1,4-glucanase (XEG) by XEG-protein inhibitor. J. Biol. Chem. May 25;287(22):18710-6. doi: 10.1074/jbc.M112.350520.
I hope you will find these pieces interesting. Your vote and follow help me open up the exciting world of plant biology and pathology to you.
Thank you @foundation for this fantastic SteemSTEM gif

impressive to say the least ! thanks for sharing ;)
you are welcome @macfarhat. Hope you are enjoying my work.