Naturally occurring transgenic crops? FUNKY Facts (1)
Naturally transgenic crops? Do they exist and are we really eating them?!?
In this post, I will describe work (Matveeva et al., 2012 & Kyndt et al., 2015) that describes the discovery and characterization of a crop plant that carries genetic elements from the plant pathogenic bacterium Agrobacterium tumefaciens. @dysprosium referred to this work in a comment on one of my other posts (on transgenic crops) for which I am thankful.
High-throughput sequencing has allowed us to catalog in great detail, the gene repertoires that populate the genomes of an increasing number of organisms. Plants are no exception to this. Because of their relevance to food production, the elucidation of crop plant genome sequences has been a principal objective. To understand how a particular genome is used or employed to create an organism, however, other sequencing-based approaches exist. These so-called Next Generation Sequencing (NGS) approaches not only allow us to define the gene sequences that are present in a given organism, but it also permits us to assess their levels of expression. Sequencing and quantification of mRNA molecules (RNASeq) can tell us about their expression patterns and provide hints about their possible function. Sequencing of small RNA molecules (sRNASeq), in turn, help us determine how the levels of specific mRNAs could be regulated. These sequencing-based approaches not only help us identify and characterize genes in ever greater detail but the vast number of sequences that are generated also enable powerful computational analyses that help understand the evolution of genomes or genes and enhance our understanding of protein function.
Now, back to this paper. To understand gene regulation in cultivated Ipomoea batatas (sweet potato), a small RNA sequencing project was initiated. One way of analyzing sequence data is to look for similarities between the sequences that have been generated and those that are already out there in databases and come from a wide range of organisms. These computationally-driven searches led to the discovery that there are small RNA molecules that have high sequence similarity to proteins, normally found in Agrobacterium rhizogenes (! This strongly suggested that a sweet potato plant had received these pieces of DNA from this bacterium. As it happens, this A. rhizogenes is well known for its ability to transform plant roots in nature, raising the possibility that at some stage, this bacterium transferred some of its genes into its host (sweet potato) and that these genes were subsequently retained in the genome.
This, of course, is one hypothesis. An alternative scenario is that the samples used for sequencing were contaminated with this soil-dwelling bacterium. It is also possible that these researchers happened to sequence a plant that was generated by transformation (in the laboratory). To test these hypotheses, the authors first determined where the transgenes were located in the plant genome. A primer walking approach was used to confirm the presence of the insert in the genome while affirming the exact location of insertion.
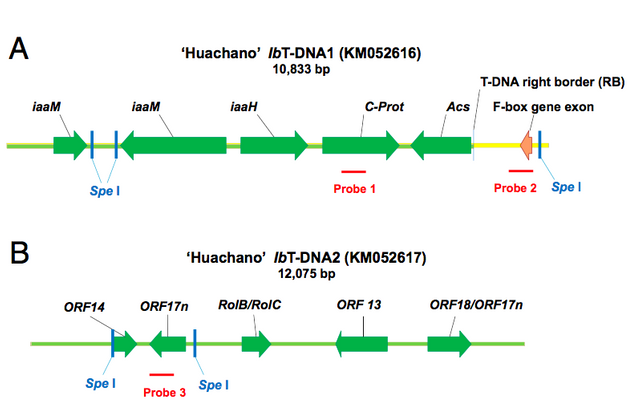
Figure 1. schematic representation of the T-DNA insertions into the Sweet potato genome. These results show that the pieces of bacterial DNA are actually taken up into the plant genome. Figure comes from Kyndt et al., (2015). Source
To rule out a unique transformation event, the authors then tested a diverse collection of sweet potato relatives for the presence of this piece of bacterial DNA. If this is a single event, only the original plant line would have the bacterial piece of DNA. If this has happened on multiple occasions or a very long time ago (before divergence and speciation events that led to distinct plant lines)the inserted sequence would be expected to be widespread. To test this, a library of clones that carry large inserts (a bacterial arteficial chromosome or BAC) and which was made from another sweet potato line, was screened to identify fragments that contain the identified DNA insertion site. Sequencing of these large clones indeed confirmed the presence of one of the fragments, although some differences were found between them.
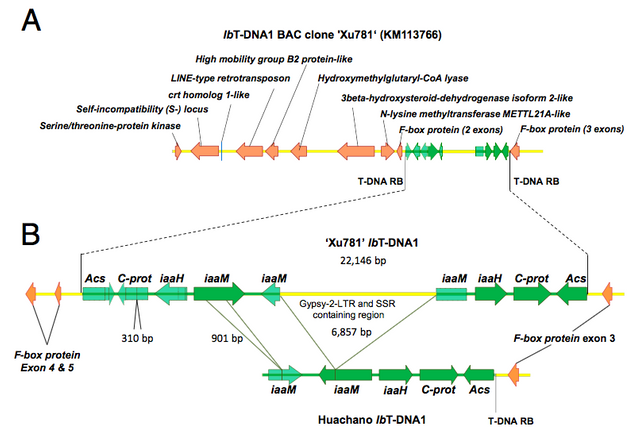
Figure 2. Comparison of DNA insertion sites between the original “Huachano” and the “Xu781” plant lines. Figure taken from Kyndt et al. (2015). Source
The authors then assessed a wide range of plant relatives to see whether these DNA fragments from A. rhizogenes were widespread within the genus. These PCR based analyses confirmed the presence of this insertion in many of the sweet potato relatives. This in turn, demonstrated that transfer of this A.rhizogenes DNA happened before the various plant species diverged from eachother. Those findings allowed the authors to conclude that this transformation event (some also call this Horizontal gene transfer) happened at a time when humans started cultivating and domesticating crops. In fact, it could even be that the actual transformation event, which led to the acquisition of new gene function and the loss of one, led to the formation of traits that were selected for in the domestication process. In other words, this could mean that without this event, man would not have adopted this plant as a crop!.
**Why is this important?**
There is an irony in the results of this study. the generation of transgenic crops is widely opposed on the basis of health and environmental concerns. What this study demonstrates is that we as a species, have been eating transgenic crops for ages (there will be more such examples) and that without transgenesis or horizontal gene transfer, our selection of edible crops would be limited.
**REFERENCES**
Kyndt, T., Quispe, D., Zhai, H., Jarret, R., Ghislain, M., Liu, Q., ... & Kreuze, J. F. (2015). The genome of cultivated sweet potato contains Agrobacterium T-DNAs with expressed genes: an example of a naturally transgenic food crop. Proceedings of the National Academy of Sciences, 112(18), 5844-5849.
Matveeva, T. V., Bogomaz, D. I., Pavlova, O. A., Nester, E. W., & Lutova, L. A. (2012). Horizontal gene transfer from genus Agrobacterium to the plant Linaria in nature. Molecular plant-microbe interactions, 25(12), 1542-1551.
I hope you will find this piece interesting. Your vote and follow help me open up the exciting world of plant biology and pathology to you.
Thank you @foundation for this fantastic SteemSTEM gif

Nice post. Horizontal gene transfer happens all the time, but its especially interesting when it happens between kingdoms. Here, between a bacteria and a plant. Keep up the good work!
Thank you!
This post has caught the eye of @MuxxyBot and has been nominated by the curation team.
If chosen it will feature in a curation post by @MuxxyBot.
An image from your post may be featured.
Fantastic, thank you!
Congratulations @huitemae! You have completed some achievement on Steemit and have been rewarded with new badge(s) :
Click on any badge to view your own Board of Honor on SteemitBoard.
For more information about SteemitBoard, click here
If you no longer want to receive notifications, reply to this comment with the word
STOPCongratulations. This post is featured in today's Muxxybot Curation Post.https://steemit.com/curation/@muxxybot/muxxybot-curation-36