Carbohydrate Metabolism in Plants - Overview
One of the main challenges I had when teaching Plant Physiology to undergraduates at Purdue University, was explaining, in relatively simple terms, how carbohydrates produced during photosynthesis are used to make starch, sucrose, cellulose, lipids and derive energy (in the form of ATP and NADH) during glycolysis and mitochondrial respiration. In particular, how is this all regulated? How do plants "decide" where to divert all of this carbon fixed during photosynthesis? I would here like to give an overview of these pathways and their regulation and point out that the concentration of phosphate (Pi) is at the heart of this regulation.
Shown below is a figure that captures the main features of these pathways and their regulation:
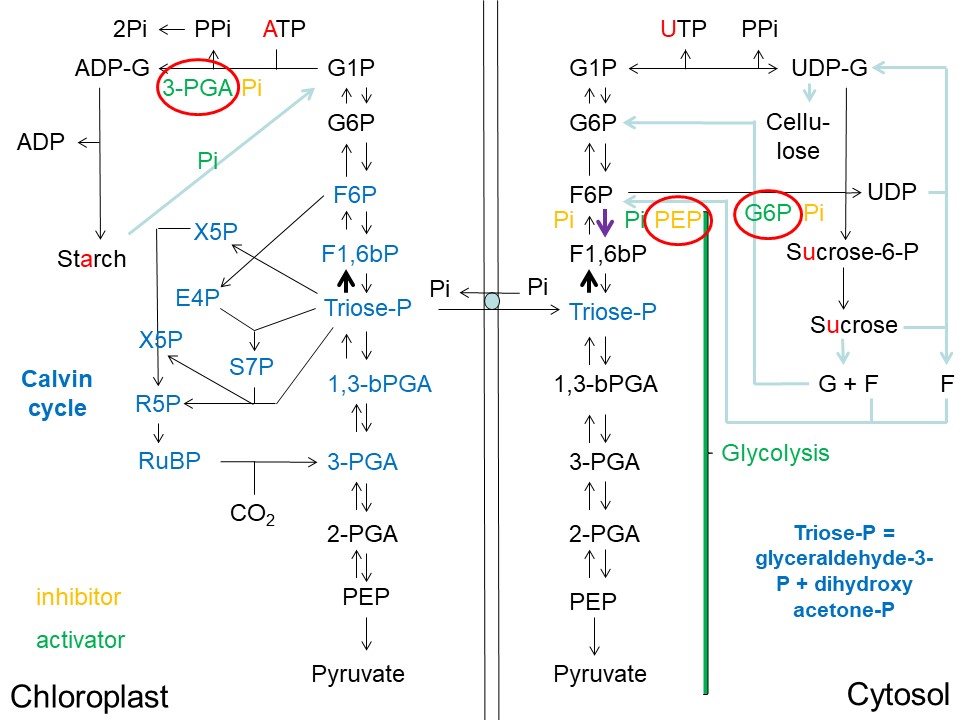
Overview of Starch, Sucrose and Cellulose Synthesis in Plants and its Regulatory Architecture (D. Rhodes)
Abbreviations used: ADP = adenosine-diphosphate; ADP-G = ADP-glucose; ATP = adenosine-triphosphate; E4P = erythrose-4-phosphate; F = fructose; F1,6bP = fructose-1,6-bisphosphate; F6P = fructose-6-phosphate; G = glucose; G6P = glucose-6-phosphate; G1P = glucose-1-phosphate; Pi = inorganic phosphate; PPi = pyrophosphate; 3-PGA = 3-phosphoglycerate; 2-PGA = 2-phosphoglycerate; PEP = phosphoenolpyruvate; 1,3-bPGA = 1,3-bisphosphoglycerate; RuBP = ribulose-1,5-bisphosphate; R5P = ribulose-5-phosphate; S7P = sedoheptulose-7-phosphate; sucrose-P = sucrose-phosphate; triose-phosphate = glyceraldehyde-3-phosphate + dihydroxyacetone phosphate; UDP = uridine-diphosphate; UDP-G = UDP-glucose; UTP = uridine-triphosphate; X5P = xylulose-5-phosphate.
On the left we have the chloroplast stroma, containing the Calvin cycle enzymes (The Calvin Cycle of C3 Photosynthesis) utilizing ATP and NADPH generated during the light reactions of photosynthesis (Light Reactions of Photosynthesis). The Calvin cycle intermediates are shown in blue.
On the right we have the cytosol (cytoplasm). Many of the reactions found in the chloroplast are duplicated in the cytosol (see vertical list of intermediates from pyruvate (at the bottom) all the way to glucose-1-phosphate (G1P) (at the top). Between the two compartments we have the inner chloroplast membrane, and the key "gate-keeper" (depicted as a pale blue circle between the two vertical lines separating the chloroplast from the cytosol), the triose-phosphate/phosphate translocator, which allows phosphorylated 3-carbon units to exit the chloroplast only in exchange for inorganic phosphate (Pi).
The upper-left quadrant leads to starch synthesis, the upper-right quadrant leads to sucrose and cellulose synthesis, the lower-left quadrant leads to pyruvate and ultimately to acetyl-CoA and lipid synthesis in the chloroplast (not shown), and the lower-right quadrant leads to glycolysis, pyruvate and ultimately to acetyl-CoA and mitochondrial respiration via the citric acid cycle (not shown).
For simplicity the names of the enzymes catalyzing the various reactions are omitted in this figure, but will be discussed in subsequent articles focusing on different quadrants. Instead I have simply listed the compounds that are known to activate (green) or inhibit (yellow) the various enzymes of these pathways. Notably, phosphate (Pi) plays a key role in regulating export of triose-phosphate from the chloroplast, as an activator of glycolysis, and inhibitor of sucrose biosynthesis, as an inhibitor of starch synthesis, and an activator of starch degradation. The various branches of these pathways are not only controlled by Pi concentration but also by pathway-specific metabolites. Thus, 3-PGA is an activator of starch synthesis. PEP is an inhibitor of glycolysis, and G6P is an activator of sucrose biosynthesis (see red circles).
Note that ATP is required for starch synthesis, while UTP is required for sucrose synthesis. This becomes easier to remember if you simply look at the first vowel in the words "stArch" and "sUcrose" (A = ATP, U = UTP).
ATP is produced from ADP + Pi in the chloroplast stroma by ATP synthase using a proton gradient generated in the light reactions of photosynthesis (Light Reactions of Photosynthesis).
ATP is produced from ADP + Pi in the cytoplasm by enzymes of glycolysis, and from ADP + Pi by ATP synthase in the mitochondrion using NADH and a proton generated during oxygen-dependent respiration of carbohydrates via the citric acid cycle. The mitochondrial-derived ATP has ready access to the cytoplasm.
It is useful to think about the relative levels of ATP and ADP + Pi in the two separate compartments, chloroplast and cytoplasm, and to do so using a "gas tank" analogy:
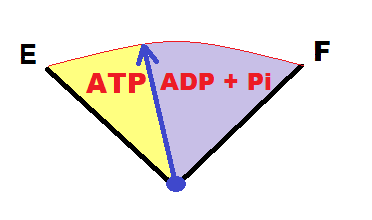
When ATP levels are high, ADP + Pi levels will tend to be low, when ATP levels are low, ADP + Pi levels will tend to be high. Essentially, the Pi concentration reflects how empty the "ATP gas tank" is.
Let's consider the scenario where both the cytoplasm and chloroplast have high levels of ATP, and low Pi levels. Such a scenario would likely occur in a healthy leaf actively photosynthesizing in the light. Triose-phosphate would tend to be retained in the chloroplast under these conditions, and diverted towards starch biosynthesis.
However, when the light is turned off, the light reactions of photosynthesis would immediately cease, ATP levels in the chloroplast would decline rapidly as ADP + Pi levels concomitantly increase. The increased Pi (and reduced levels of 3PGA) in the chloroplast would inhibit starch synthesis, and the elevated Pi levels would activate starch catabolism!
If Pi levels rose in the cytoplasm, perhaps due to inadequate supply of carbohydrates for mitochondrial respiration and ATP production, triose-phosphate export from the chloroplast would be accelerated and the elevated cytoplasmic Pi concentration would tend to divert this carbon towards glycolysis (and ultimately to mitochondrial respiration via the citric acid cycle) and away from sucrose and cellulose biosynthesis.
I would encourage readers to consult the recent articles by @kingabesh on CHEMISTRY AND METABOLISM OF BIOMOLECULES #1: INTRODUCTION (CARBOHYDRATES: MONOSACCHARIDES AND DISACCHARIDES) and CHEMISTRY AND METABOLISM OF BIOMOLECULES #2: INTRODUCTION (CARBOHYDRATES: OLIGOSACCHARIDES AND POLYSACCHARIDES) for the structures of sucrose, starch and cellulose.
Please feel free to comment or ask questions below. I will try to answer as soon as possible.
Interesting and quality post as usual, which I will read more than once. Carbohydrate metabolism and biochemistry is an area in which I have less experience than DNA, RNA, proteins, and lipids. I enjoy trying to fill in my gaps
Thank you!
Your overview of carbohydrate metabolism in plants is top notch. It’s sad to see your students had a hard time understanding plant physiology under your tutorship when you explain these processes so succinctly and clearly. Looking forward to the different fragments of this article.
Thanks for the mention too ! :)
Thank you!
I feel like my eyes is being opened to a whole new world of carbohydrate metabolism. I can say for sure there can be no clearer explanation than this. Thank you david, and please, more.
Thank you, I hope to have another one posted tomorrow.