DNA METABOLISM: REPLICATION OF DNA.
INTRODUCTION
As the site of genetic information, DNA occupies a central position among biological macro molecules.The DNA nucleotide sequence encodes the primary structure of all cellular RNA's and proteins and through enzymes, indirectly affecting the synthesis of all other cellular components.
The metabolism of DNA consist of a set of enzyme catalyzed processes that mainly involve three stages:
- Replication
- Repair
- Recombination.
The enzymes that synthesize DNA may copy DNA molecules that contain million of bases. The essence of DNA copying or Replication is so that the two daughter cells produced at the end of cell division would have the same genetic information as their parent.
This is solely DNA Replication.
The double helix structure of DNA provides a simple mechanism for replication. The two strands are separated and then each strand complementary DNA is recreated by an enzyme called DNA POLYMERASE.
The DNA polymerase was first synthesized from E. coli cells.

A single polypeptide enzyme called DNA POLYMERASE I. Further research led to the discovery of four other DNA Polymerases. The main function of this enzyme is to synthesize DNA from nucleotides which are the building blocks of DNA. The several forms of the enzyme works in pairs to copy one molecule of of double stranded DNA into two new double stranded molecules. It is required to replicate or duplicate the cells DNA, so that the original DNA molecule can be passed to each daughter cells thereby transmitting genetic information through generations.
The reaction carried out by this enzyme is a phosphoryl group transfer. The nucleophile is the 3'- hydroxyl group of the nucleotide at the 3' end of the growing strand. The site of nucleophilic attack is the alpha phosphorus of the incoming deoxynucleoside 5' triphosphate. Inorganic pyrophosphate is released in the reaction.
EQUATION OF REACTION.
(dNMP)n + dNTP = (dNMP)n+1 + PPi
Catalysis by DNA polymerases prominently involves two magnesium ions at the active site. one helps to deprotonate the 3'- hydroxyl group making it a more effective nucleophile and the other binds to the incoming dNTP and facilitates departure of the pyrophosphate. The active site of this enzyme DNA POLYMERASE I accomodates only base pairs. An incorrect nucleophile may be able to hydrogen-bond with a base in the template, but it generally will not fit into the active site.Incorrect bases are rejected before the phosphodiester bond is formed.
One mechanism intrinsic to DNA polymerase is a separate 3'-5' exonuclease activity that double checks each nucleotide after it is added. This nuclease activity permits the enzyme to remove newly added nucleotide and is highly specific for mismatched base pairs. Translocation of the enzyme to the position where the next nucleotide is added is inhibited and this occurs if the polymerase has added the wrong nucleotide. The 3'-5' exonuclease activity removes the mispaired nucleotide and the polymerase begins again. This activity is known as PROOF READING.
DNA REPLICATION.
The concept of Template was used to explain the ability of organisms to replicate themselves. This template is a structure that allows molecules to be lined up in a specific order and joined to create a macro-molecule with a specific sequence and function. The discovery of the Watson and Crick model of DNA further explained how DNA could act as a template for the replication and transmission of genetic information.
As earlier understood, the Watson and Crick's DNA model is that of a double helix in which one strand complements the other. Upon unwinding, each strand acts as a template for the replication of the other.
Replication proceeds with a degree of fidelity and are of two types:-
CONSERVATIVE and
SEMI- CONSERVATIVE.
The conservative replication is one in which the parental strands never completely separate. Therefore, after one round of replication, one daughter duplex contains only parental strands and the other daughter strands.
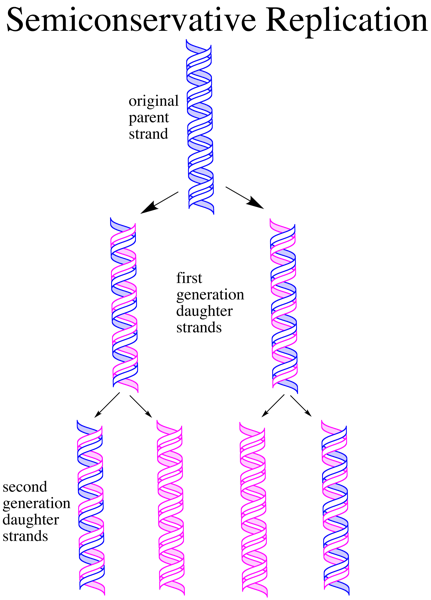
The semi conservative on the other hand, each daughter duplex consists of the parental strand and a newly synthesized strand formed from the complementary strand.
A typical example is replication in E. coli.
Meselon- Stahl expeiment.
- Cells were grown in a medium containing only nitrogen for many generations. All the nitrogen in their DNA was shown by a single band when centrifuged in CsCL density gradient.
- The cells were transferred to a medium containing only light nitrogen, cellular DNA was isolated after one generation attained a higher position in the density gradient.
- A second cycle of replication yielded a hybrid DNA band and another band containing only DNA therefore confirming that semi conservative replication took place in E. coli cells.
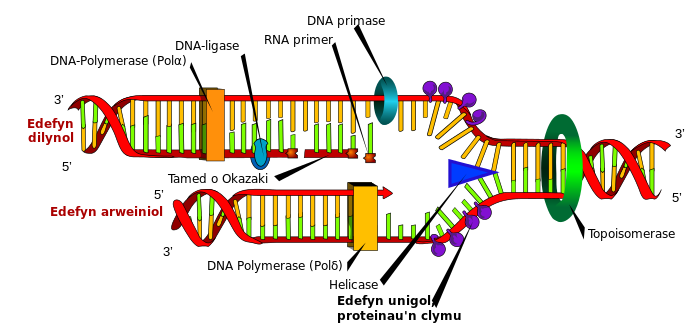
EVENTS IN DNA REPLICATION.
American enzymologist and nobel prize winner Arthur Kornberg compared DNA replication to a tape recording of instructions for performing a specific task. He said and I quote ;
"exact copies can be made from it, as well as the tape recording so that this information can be used again and else where in time and space."
In reality, the replication process is far more complex and has been categorized into five stages:
IDENTIFICATION OF SITE OF THE ORIGIN OF REPLICATION.
There are specific sites for the origin of replication. At these sites there is an association of sequence specific DNA binding proteins with a series of direct repeat DNA sequences.
Adjacent to this site of origin is a A + T rich region. When a protein interacts specifically to the site of origin of replication. The result is the denaturing and unwinding of the adjacent A + T rich region.
UNWINDING OF DNA TO FORM ssDNA THAT ACTS AS A TEMPLATE.
Protein interaction with the site of origin of interaction delineates the site and produces a region of short ssDNA that acts as a template for initiation and synthesis of nascent DNA strand. The major enzyme here is the DNA helicase which allows for processive unwinding of DNA.
single strand binding proteins bind to single strand DNA strand, stabilizing the complex and preventing re annealing.
FORMATION OF THE REPLICATION FORK.
The replication fork is formed following this order of sequence:-
- DNA helicase unwinds a short segment of the dsDNA.
- A primase initiates synthesis of RNA molecule that is essential for priming DNA synthesis.
- The DNA polymerase initiates nascent daughter strand synthesis and
- ssB proteins binds to ssDNA to prevent premature reannealing of ssDNA to dsDNA
INITIATION AND ELONGATION OF DNA.
The priming process comes before the elongation and it involves nucleophilic attack by the 3'- OH group of the RNA primer on the alpha phosphate of the first entering deoxynucleotide triphosphate with the release of pyrophosphate.
In the elongation process the 3'- OH group of the newly attached deoxyribonucleotide monophosphate is then free to carry out nucleophilic attack on the next entering deoxyribonucleotide triphosphate, at its alpha phosphate group with the release of PPi.
Note that the nascent DNA is always synthesized in the 5'-3' direction because DNA polymerases can add a nucleotide only to the 3' end of a DNA strand.
The selection of a new nucleotide entrant is governed by proper base pairing rule.
FORMATION OF REPLICATION BUBBLES AND LIGATION OF NEWLY SYNTHESIZED DNA MOLECULES.
The period of genome replication is approximately 9 hours in mammals and the average period required for tetraploid genome formation from diploid is approximately 150 hours if done from a single site of origin of replication.
However, replication occurs bidirectionally and from multiple sites of origin, thus it occurs in both directions in ll chromosome and both strands are replicated simultaneously resulting in the formation of replication bubbles.
In mammals after many Okazaki fragments are formed, the replication complex begins to remove the RNA primers to fill in the gaps by their removal with the proper base paired deoxynuclotides, and then to seal the fragments of newly synthesized DNA by an enzyme that requires energy from ATP. This neak sealing enzyme is called DNA LIGASE.
Replication of DNA is a highly coordinated process in which the parent strand are unwound and replicated simultaneously. The replication forks originates at unique points in the DNA molecule and this was determined by a technique called DENATURATION MAPPING. This was developed by Ross Inman and some of his colleagues. Using the 48,502 bp chromosome of bacteriophage, he showed that DNA could be denatured at sequences unusually rich in A=T base pairs, generating a reproducibile pattern of single stranded bubbles. Isolated DNA containing replication loops can be partially denatured in the same manner allowing for the position and progress of the forks to be measured and mapped using denatured regions as points of refrence.
For circular DNA molecules, the two replication forks meet at a point on the side of the circle opposite the origin.
- Replication of DNA occurs with very high fidelity and at a designated time in the cell cycle. It is semi conservative, each strand acting as a template for the new daughter strand. It is carried out in three phases: initiation, elongation and termination. The process starts at a single origin in bacteria and usually proceeds bidirectionally.
- DNA is synthesized in the 5' -3' direction by DNA polymerases. At replication fork, the leading strand is synthesized continuously in the same direction as the movement of the fork. The lagging strand is synthesized discontinuously as Okazaki fragments, which are also ligated.
- The fidelity of DNA replication is maintained by base selection by the polymerases, specific repair systems for mismatches left behind after replication, and a 3'-5' proofreading exonuclease activity that is aprt of most DNA polymerases.
- Most cells have several DNA polymerases. The DNA polymerase I is responsible for special functions during replication, recombination and repair.
- The separate initiation, elongation and termination phases of DNA replication involves an array of enzymes and protein factors, many belonging to the ATpase family.
REFERENCES.
- Lehninger Principles Of Biochemistry, seventh edition by David L. Nelson and Michael M. Cox.
- MN Chatterjea Textbook Of Medical BIochemisty, eighth edition.
- conservative replication
- molecular events of replication
Resteemed your article. This article was resteemed because you are part of the New Steemians project. You can learn more about it here: https://steemit.com/introduceyourself/@gaman/new-steemians-project-launch
Congratulations @bundis10! You received a personal award!
Click here to view your Board of Honor
Congratulations @bundis10! You received a personal award!
You can view your badges on your Steem Board and compare to others on the Steem Ranking
Vote for @Steemitboard as a witness to get one more award and increased upvotes!