The solution to my “Decode the message” challenge lies within the Nobel Prize for Medicine/Physiology from 1968.
Hello fellow Steemiacs,
in this post I will give you the solution to my previous “Decode the message” challenge and announce its winners!

Pixabay
What's up with the thumbnail?
To briefly recapitulate - the task was to decode this message from the bottle:

If you look at the letter composition of the text, you will notice that there are only four distinct letters in it:
While you can look for specific patterns, you won't find any, except that the letter H is the least and U the most frequently occurring.
The question is how a combination of four letters can code for a message similar to the one that I was looking to code myself (“WHEN WILL ETH DIP”).
A four-letter code might or might not immediately ring a bell. However, if you apply the most simple cryptographic method of shifting the letters along their order in the alphabet, you will come up with a transformation that takes each letter one step back - substitute B with A, D with C, H with G, and U with T.
A, C, G and T (have you watched the movie Gattaca?) stand for the four bases that make up the store of information that is the most unifying feature of all life on Earth – the DNA (deoxyribonucleic acid).
So, after performing the first decryption step, the message looks like this:
The two winners of the contest have successfully resolved the encryption because they knew how to read the genetic code. In addition, they also knew how to write in the genetic code themselves, and encrypted my message “WHEN WILL ETH DIP”. Their answers for the encryption were:
and
UDBDUBUUBBHHUBUBUDUUBBUHHHUUUDUUBBBHBBHUBUDDBUUBBUUUUDBUHDDBUUBDUBUDB
As you see, these two messages, while bearing some resemblance, are not quite the same. Read on to find out why they are, nevertheless, both correct and what the original message is saying!
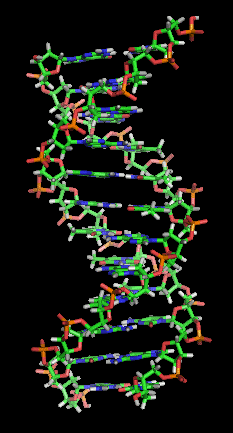
To understand the essence of the genetic code, we need to go back to 1953, when Watson & Crick proposed the double-helix structure of DNA.
The DNA is a double-stranded molecule with two single-strands winding themselves around each other in an antiparallel manner resulting in a helical structure, as a whole resembling a corkscrew stairs.
By Zephyris at the English language Wikipedia, CC BY-SA 3.0, Link
Antiparallel means that one strand is oriented in a sense direction (denoted as 5'->3', referring to the chemical position of phosphate groups) while the second strand is oriented in the antisense (3'->5') direction.
DNA is a very stable molecule, but how do the two strands stick to each other? This is where the true beauty of the DNA molecule reveals itself.
There are two structurally related kinds of bases: the purines A(denine) and G(uanine) and the pyrimidines C(ytosine) and T(hymidine). In the spatial orientation that these bases find themselves within the double-helix, a chemically stable state can only be reached when purines on the sense strand pair up with pyrimidines on the antisense strand, and vice versa.
Thus, A pairs up with T (A:T), and G pairs up with C (G:C). The pairing up is established by the formation of so-called hydrogen-bonds (signified by “:”). This is a kind of chemical bond that, for example, allows frozen water to form ice crystals, or, makes ethanol a good solvent.
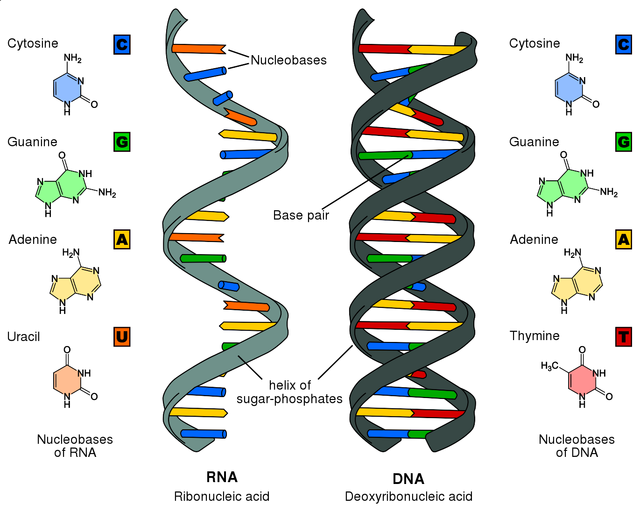
Structure of DNA and RNA. Refer to this image later in the post when we come to RNA molecules.
By Sponk- chemical structures of nucleobases by Roland1952, CC BY-SA 3.0, Link
Because of this pairing principle, the two strands are complementary to each other. Knowing the sequence of bases in the one strand you automatically know the sequence of bases in the other strand.
For instance, if the sense strand reads: 5'-CCTAGT-3', then the antisense strand must read as 3'-GGATCA-5' , or by convention, in reverse complementary, 5'-ACTAGG-3'. See how we got there?
Did you know that the DNA in the human genome, if spelled out, would fill up an equivalent of a thousand book copies of the bible?
Now that we know the arithmetics of DNA, we need to understand what the output of the stored information is and which algorithm governs its translation.
By the 1950ies, it was already known that DNA carries discrete units, called genes, that can determine a phenotype, i.e. a biological feature such as the color of a pea.
According to the one-gene-one-enzyme hypothesis, put forth by Beadle and Tatum in 1941, each gene codes for the production of one specific protein. (Enzymes are proteins, but not all proteins are enzymes.) One such enzyme is the alcohol dehydrogenase, that helps sober you up after a night out.
Proteins are made up of twenty chemically distinct species of amino acids. Each of the twenty amino acids is abbreviated by one or three letters, depending on the application. For example, Glycine is abbreviated by Gly or G. If you are into bodybulding, you must have heard of Glutamine, which is abbreviated by Gln or Q. Phenylalanine is Phe or F. And so on.
In total, there are twenty distinct letters in the alphabet that are represented by amino acids. I hope you can see now, how the four-letter genetic code might be read as a sequence of a distinct set of twenty letters.
While in December 1968, Apollo 8 took off onto its mission to orbit around the moon, on Earth, three gentlemen, Robert W. Holley, Har Gobind Khorana, and Marshall W. Nirenberg, were honored with the Nobel Prize for Medicine/Physiology, for the perhaps most important and ground-breaking discovery in biology - the working principle of the genetic code

The Nobel laureates of the Medicine/Physiology Nobel award 1968.
Source
From DNA to protein, there are several additional molecules and molecular complexes involved that we briefly need to look into, to understand what the Nobel laureates did to deserve this highest scientific award.
DNA needs to be kept safe to allow its accurate inheritance to daughter cells. Furthermore, cells require certain proteins in very large amounts. Therefore, the DNA needs a host of messengers to translate its genes into functional products. This function is assumed by the so-called messenger RNA (mRNA), a ribonucleic acid that is chemically and structurally very similar to DNA, but is single-stranded and due to its chemical nature has a U(racil) base instead of T. U and T are equivalent when it comes to their coding function: they both pair up with A.
Once produced from its DNA gene template, the mRNA docks onto the ribosome, a molecular machinery that synthesizes proteins by reading the mRNA code.
The ribosome needs adapter molecules to correctly read the message. These adapter molecules were identified to be transfer RNAs (tRNAs). Distinct tRNAs are loaded with distinct amino acids by their respective aminoacyl-tRNA synthetase enzymes.
Therefore, tRNAs are the central decoding elements linking the genetic code to amino acids.
The Nobel laureates and other biochemists, chemists and biologists all contributed to cracking the genetic code by accomplishing the following milestones:
- An in vitro code translation system with Escherichia coli (a common intestinal bacterium) extracts was devised which enabled the scientists to provide defined factors, such a RNA molecules and radioactively labeled amino acids, and observe how and to what results they interact with each other biochemically.
- Biochemists figured out how to synthesize RNA templates with defined sequences of ribonucleotides (i.e. bases within the RNA strand)
- The hypothesis was that the genetic code is read in distinct units that each correspond to one of the twenty amino acids. Given that there are four different bases, the minimal coding units must be at least triplets (e.g. ATC), because singlets would only be able to encode four different amino acids (=4), and doublets only sixteen (=4²).
There are 64 (=4³) possible triplet base combinations, thus, being theoretically able to cover all twenty amino acids.
By providing either single-, double-, triple-, quadruple-base etc. (i.e. A, AA, AAA, AAAA, ...) molecules of RNA they observed that only when there were at least a triple sequence of A, the E. coli translation system was able to bind the amino acid lysine. With hextuples (AAAAAA), but not with quadruples or quintuples, the binding was higher than with triplets.
This was very simple, but compelling evidence, that the genetic code is read in triplets.
You can probably guess by now, how the scientists figured out the genetic code. They threw all possible, repetive base sequence combinations and observed which amino acid would bind to the E. coli translation system.
I.e. after providing polyU-RNA molecules (strings of sequential Uracile bases), they obtained peptides consisting of solely the amino acid Phenylalanine. They put in polyUC RNA molecules (resulting in two repeating triplets UCU and CUC) and observed binding of exactly two amino acids - serine and leucine). They continued with repeating units of up to four bases. From there, they could do the combinatorial math and came up with the following genetic code, consisting of 64 triplets or codons:
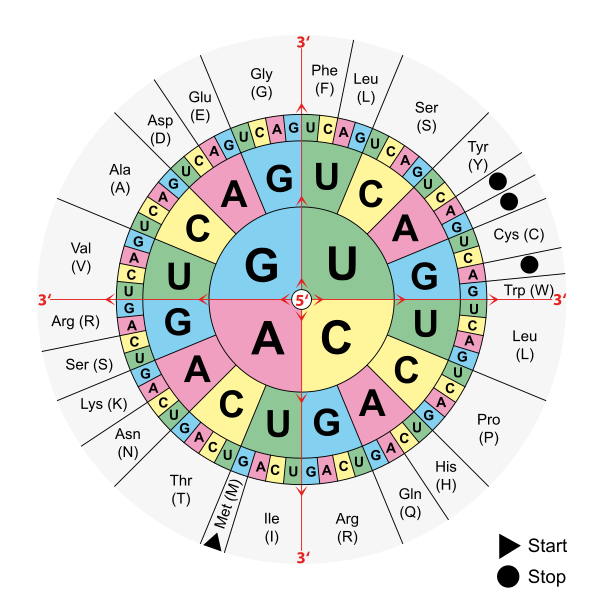
This depiction of the genetic code is to be read from inwards of the circle (5') to outwards (3'). You can use this "code sun" to decode my message. Follow the instruction further below. - It was found that one codon (ATG), the start codon, is reserved for the initiation of a protein chain and at the same time codes for the amino acid methionine.
Three codons signal the termination of protein synthesis and are called stop codons. - The code was found to be essentially the same in bacteria, amphibae and mammalian cells, therefore it is referred as the standard genetic code.
Even though there are minor deviations, it is so similar that you can take the Pax6 gene from the mouse, put into a fruit fly and observe how an eye is formed (Pax6 being the intructive gene for eye development in many organisms). - One of the final proofs for the triplet-amino acid relationship was the sequencing of the adapter transfer RNA, that carries the amino acid alanine. This was, indeed, the first ever gene completely sequenced, which was a tremendous undertaking at that time!
The alanine-tRNA carries, at a structurally exposed position, the complementary base sequence – called the anticodon - to that of the known alanine codon on RNA (and thus the sense DNA strand).
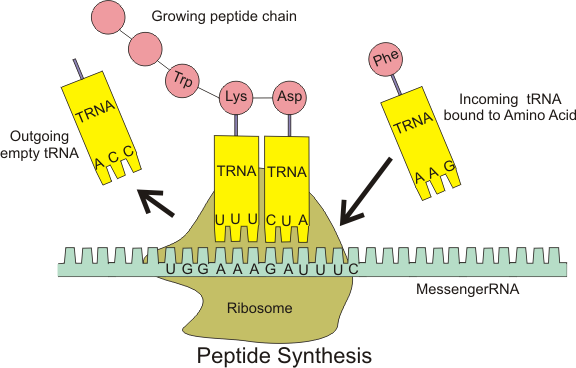
A summarizing depiction of the translation principle.
By Boumphreyfr (own work), CC BY-SA 3.0 , via Wikimedia Commons"
As you have noticed, there are three times more codons than amino acids, resulting in several codons encoding the same amino acid. The genetic code is, therefore, called degenerate.
This is the reason why I got two differently encrypted messages from the contest winners, which nevertheless, carried the same message!
Finally, after this long read, we come to the solution of the original message. There are two problems left, now that we know the base sequence:
1. Does this sequence correspond to the sense or antisense strand?
2. In which frame is it to be read, i.e. do we start reading from the first, second or the third letter of the sequence?
I will offer you a very simple solution to this problem: a bioinformatic tool.
You can go to https://www.ebi.ac.uk/Tools/st/emboss_transeq/ , paste the sequence from above, and select “6 (All six frames) in the “FRAME” pop up window.
Click “Submit” and after a short waiting period you get the result!

Now you have to select which of the six possible translations makes most sense. Of course it is this one:
Each * results from a stop codon. The message was encoded in the antisense strand. Therefore, without using this bioinformatic tool, you would first need to obtain the reverse complementary, sense strand, and start translating according to the genetic code from there.
The biggsest problem with this encryption method is that six letters of the alphabet are not represented by the amino acid single letter code (namely B, J, O, U, X and Z). This can be alleviated, however, as there are a lot of degenerate codons left to be reassigned to new letters.
Finally, I am announcing the winners of the challenge! There were only two participants, but fortunately for them, they could split the 3rd prize among them!
The 1st prize of 6.102 SBDs went to @tking77798 !
The 2nd prize of 4.068 SBDs went to @eonwarped !
Congratulations and thank you for the participation! I am really happy that you managed to read the gibberish message!
To all others, let's spread the message: MY STEEM IS CASH!
One last fun fact: Very recently, scientists introduced an artificial base into the genome of a bacterium that can be translated into a synthetic amino acid! This involved the introduction of a specialized tRNA and its respective aminoacyl-tRNA synthetase! Thus, the genetic code is not written in stone and can be expanded artificially which might, in future, e.g. help to synthesize certain drugs.
Yours, @replichara
Sources and further reading:
- Robert W. Holley, Nobel lecture, December 12, 1968.
- H. Gobind Khorana, Nobel lecture, December 12, 1968.
- Marshall Nirenberg, Nobel lecture, December 12, 1968.
- Koonin E.V. and Novozhilov A.S. Origin and evolution of the genetic code: the universal enigma. IUMBM Life. 2009 Feb; 61(2): 99-111.
- Alberts B., Johnson A., Lewis J., Morgan D., Raff M., Roberts K. and Walter P. Molecular Biology of the cell. 6th Edition. 2015, Garland Science.
- @irime 's blog
It was a fun puzzle and the DNA focus made it great. Thanks @replichara!
Such an interesting challenge! I would have never guessed, ha ha. Super impressed with the two who did though.
I wasn't sure if anyone would manage, but I thought that at least someone with a biology education would be able to connect the lines :-) And it proved to be the case.
Craig Venter and his team produced a synthetic bacterium from scratch, and left a watermark message in its genome in 2010.
https://www.theguardian.com/science/2010/may/20/craig-venter-synthetic-life-form
And here are the translations of the messages:
https://singularityhub.com/2010/05/24/venters-newest-synthetic-bacteria-has-secret-messages-coded-in-its-dna/#sm.0000pvwks8spqeenz6u116ze918dd
Did not exactly know about this, before I made the puzzle :-)
I have a biology background... 🙈 I think you need to have the ability to think outside the box though.
Julian Savulescu, mentioned, in the first article above, that ‘Venter’ is playing God, by creating new species.... If the intention is to create organisms for the greater good, then there is no problem? God gave us a beautiful mind, capable of creation. We create music, art, film everyday. Surely ‘biological art’ is not a sin?
Julian Salvulesci is an interesting man though. I have read a lot of his work on ethics in sport. He holds some interesting opinions.
I didn't have Savulescu on my radar. Let's see, if synchronicity will lead me to stumble upon him again :-D
To me, his quote in the article, sounds rather neutral, without taking judgement. Myself, I don't see a problem with this synthetic creation. "Playing god" is a hollow term to spread FUD among people.
Humans have played god since they lighted the first fire and learned how to produce and use tools. Humans have played god when they domesticated wolfs and bred them towards ridiculous dog races.
The difference with genetic engineering is that it can be precise, goal-directed and safe, if executed smartly.
The intentions of the people behind the technology are critical. A.I. Is another technology which has so many beneficial uses, yet, is dangerous in the wrong hands.