The (Not So) Scarry Science Behind Genetic Engineering and Biotechnology
Impressive tools are now available for manipulating DNA and changing the genomes of organisms to our benefit as a species.
The fundamental mechanisms cells use to replicate and transcribe DNA provided researchers with insight into how to modify genes, how to transfer genetic material from one organism to another, and how to rapidly sequence genes. These tools provide the basis for genetic engineering and biotechnology, which are rapidly transforming our lives. We explore this strange new world by showing how genetic engineering and biotechnology have been applied in medicine and agriculture.
Our understanding of genes and their expression have led to the current revolution in genetic engineering and biotechnology.
This is a good moment to provide this overview and if you have followed my biology series thus far, we have now learned much of what we need to know to understand what genetic engineering and biotechnology are all about.
In many ways, the promise of biotechnology began with Watson and Crick’s first description of the DNA double helix, because many of the fundamental tools we use to analyze and manipulate genes today depend on the complementary base pairing rules they described. Advances in genetic engineering and biotechnology over the last several decades have had as much to do with developing techniques for conveniently or quickly working with DNA as with new conceptual understanding.

Genetic engineering involves the production of recombinant DNA.
We have seen recombinant genotypes and phenotypes in meiosis, where the term simply referred to new mixtures of traits from two different parents. Recombinant DNA has a similar meaning, with two important differences.
- When creating recombinant DNA, we determine by choice which DNA will be included. Meiotic recombination, on the other hand, works by chance.
- The second difference is that we can combine DNA from completely different organisms if needed or desired.
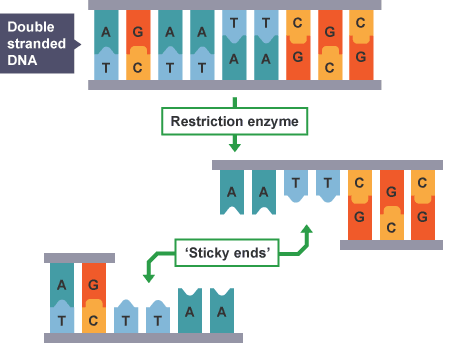
The discovery in the 1960s of a particular class of bacterial enzyme called restriction endonucleases (or restriction enzymes) was a crucial first step in learning how to manipulate genes.
Restriction enzymes are defense mechanisms bacteria use to break down the DNA of viruses, thus preventing viruses from infecting a bacterial cell. They cut DNA apart at sequences called recognition or restriction sites. Restriction sites are typically only a few bases long. The two strands at a restriction site usually are palindromic, which on DNA means that the same base sequence occurs on both strands in reverse directions (e.g., GAATTC on one strand with CTTAAG on the other). This is the case because of complementary base pairing.
The bacteria’s own DNA is protected from the action of its restriction enzymes, because in the bacteria, the sequences the enzyme recognizes are chemically modified by adding methyl groups (methylated). Because restriction enzymes cut between two particular bases in a restriction sequence, each cut piece now has a short section of single-stranded DNA extending beyond the double-stranded portion of the molecule. The exposed bases on these “sticky ends” will align through base pairing with any strand that has the appropriate complementary sequence, providing a way to recombine cut pieces of DNA. The key to creating recombinant DNA is the fact that the sticky ends of any fragments of DNA cut by the same restriction enzyme—regardless of which organism the DNA came from—will be complementary to each other.
DNA ligase, the enzyme that stitches together Okazaki fragments during DNA replication, can be used in the lab to stitch together the sticky ends of DNA pieces, creating new recombinant DNA. Hundreds of restriction enzymes have been discovered, each recognizing a different DNA sequence and, thus, each cleaving DNA at different points along the overall length of the molecule.
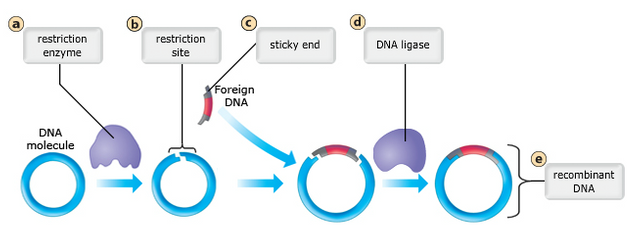
The second major advance in biotechnology was the ability to introduce recombinant DNA into a new organism.
Early work used prokaryotes as hosts for recombinant DNA, because bacteria normally transfer short pieces of DNA from one to another. Bacteria transfer DNA through conjugation, in which small pieces of DNA, called plasmids, are transferred from one cell to another.
- Restriction enzymes can cut open a plasmid and allow the insertion of a piece of genetic material that has been clipped out with the same restriction enzyme.
- The plasmid containing the added genetic material can then be inserted into new host cells using the natural process of conjugation.
- A genetically modified bacterium will use its own DNA synthesis machinery to make more copies of the recombinant gene as it divides and will use its own protein synthesis machinery to produce the protein coded for by the inserted gene.
This method works, but it has some practical problems.
- First is the difficulty of determining whether the appropriate plasmid has been created, because restriction sites occur frequently, resulting in many DNA fragments.
- It may also be difficult to identify bacteria that have incorporated the desired plasmid. One way to do this is by incorporating not only the desired gene but also an easily identified genetic marker or reporter gene.
- A major problem is that introducing eukaryotic genes into prokaryotic organisms also requires adding a prokaryotic promoter mechanism.
- In addition, eukaryotic genes have introns that must be removed. This is normally done by using mRNA and reverse transcriptase to create artificial genes that lack introns (complementary DNA or “cDNA”).
Other methods of inserting foreign DNA into both prokaryotic and eukaryotic cells have in common the use of a vector that transports DNA into cells, where it is expressed, if not incorporated into the genome.
For example, one method involves creating small artificial chromosomes with desired characteristics that function as normal chromosomes in host cells. This has been successful with yeast, and considerable progress has been made with an artificial human chromosome. Creating transgenic organisms becomes increasingly difficult as the host organism becomes more complex, but the routine insertion and expression of foreign DNA into any organism may not be far away.
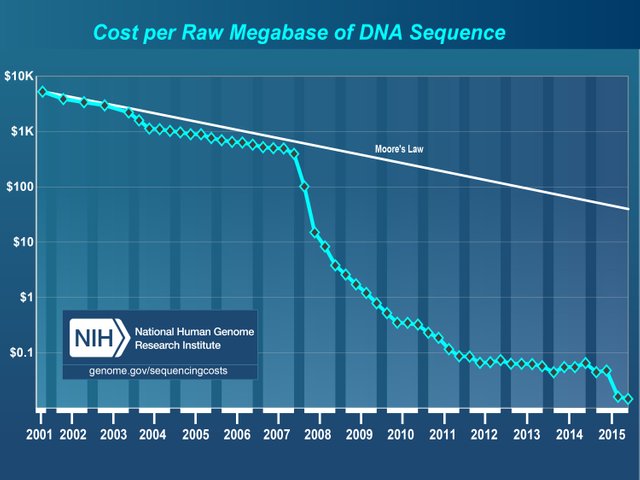
A third major advance in DNA technology has been the development of methods to determine rapidly the sequence of bases in a molecule of DNA.
The most visible consequence of the development of sequencing methods has been the ability to sequence the entire genomes of some organisms, including humans. Excluding bacteria, about a dozen genomes have been completely sequenced to date. In many cases, this kind of detailed knowledge is what makes possible much of the genetic engineering we have discussed.
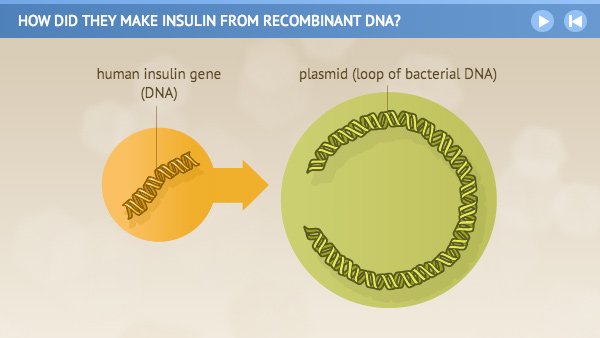
Although practical applications are still being developed, genetic engineering and biotechnology have already led to important commercial and medical benefits.
The broadest definition of biotechnology is simply the manipulation of organisms to make useful products. Selective breeding of plants and animals is, in this sense, an old form of biotechnology, but it is quite slow. The first and most widely used application of biotechnology has been to produce proteins that previously could be obtained only through tedious or costly means. Insulin used to treat diabetes could previously be harvested only from tissues taken from cows and pigs. This process is costly, it is difficult to purify the insulin, and the insulin did not function properly in some patients because the animal protein differs from the human form. Insulin is now produced commercially by bacteria that have had the human insulin gene added to their genomes, allowing them to produce the human form of the protein abundantly and relatively cheaply.

In the United States, hundreds of transgenic crop plants are in common commercial use.
Plants may be modified for insect or drought resistance, or increased nutritional value. However, many critics argue against the use of transgenic food species because of the unintended consequences gene manipulation might have or example, overproduction of pesticides or other toxic compounds or the decimation of a wild species by a genetically modified one. These possibilities are real and will need to be weighed against the benefits of transgenic organisms, especially as population pressures cause an increased demand for food.
Sources:
Image Credits:
http://www.zocalopublicsquare.org/wp-content/uploads/2016/06/Genetic-engineering-event-LEAD.jpg
http://cdn-blogs.tribune.com.pk/2014/11/24736-genetic-1417251298-120-640x480.jpg
http://www.conserve-energy-future.com/wp-content/uploads/2013/07/Genetic_Engineering.jpg
http://www.bbc.co.uk/staticarchive/919ad613beb8fe3abca51b5f283fa81f40512166.gif
https://s3.amazonaws.com/classconnection/108/flashcards/8894108/png/screen_shot_2015-11-24_at_100241_am-1513A7384336D2E4EAF.png
https://spillerlab.files.wordpress.com/2011/03/pglo11.jpg
https://lifesite-cache.s3.amazonaws.com/images/made/images/remote/https_s3.amazonaws.com/lifesite/Shutterstock/human_embryo_810_500_55_s_c1.jpg
https://www.genome.gov/images/content/costpermb2015_4.jpg
https://www.nlm.nih.gov/exhibition/fromdnatobeer/img/exhibition-recombinantDNA.jpg
http://1.bp.blogspot.com/-YKqDYDd-QWs/VXRbt88d5-I/AAAAAAAAKYI/4LcLorn6kwg/s1600/gmo-orange-kiwi.jpg

BTC: 1JQWNxziArfaCtxsP3tUQ4VSTBY9jresmt 



Upped, this article reminds me of one of my first posts in Steemit "DIY Transgenics (GMOs)". I seriously expect that you get more out of this post than I did with mine back then :)
Thank you for your continued interest in my articles @renzoarg. Let's hope it does as well as you think. My confidence could use the boost if nothing else.