Epigenetics - Not Quite Transgenerational Yet
Warning: Relatively Long, yet relatable Science Post
An analogy, to begin with, is that genes are to epigenes what hardware is to software: We can overclock and tweak its use while running it but don't expect us to pimp it for a new rig. Not even if you change the name from software to epihardware
Many know about the Siddhartha Mukherjee's fiasco in The New Yorker's article "same but different" in (May 2, 2016) about epigenetics. In case you don't, an award-winning cancer researcher did a piece on basic epigenetic science that some experts in the field consider misleading and incomplete to say the least.
I here will show you source studies, that even if old are the way to go for basic understanding. Is not mere history. ¹
Heritability and Inheritability studies
Is DNA the whole picture?
With the Human Genome Project (1990 - 2003), some questions appeared unanswered:
≤2 x 10⁴ gene coding proteins. Why so few? (Initially, researchers expected 10⁵ - 4x10⁴) ²
Bear in mind that the number is similar across species like worms, flies, humans.Where does variability comes from if you have twins? (since their DNA is identical but their phenotypes is not exactly the same)
Is the path to eradicating all diseases already in our DNA? (multifactorial diseases like coronary heart disease point to something different)
The Avery-MacLeod-Carty experiment
People once thought that proteins carry the genetic information. 1944 was the pivotal moment when we knew that the heritable components in DNA are the base of our heredity, as the bacterial transformation to pathogenic in pneumococcus pneumonia for purified extracts based on the Griffith's experiment showed.³

Soruce
There's no current evidence for this extra layer of information for transgenerational heritability (outside of plants), but there's evidence for critical periods of gene regulation in cell differentiation for some cell lines. ⁴
Canalisation
The study of developmental biology by Waddington in 1942 offered a theory for how similar phenotypes can be generated through different genetic information. The Epigenetic landscape. ⁵ To express how complex cellular patterns would be derived specifically from genes in the embryologic development.
The famous visual representation in the image is similar to the chaos theory explanation in Jurassic Park of why the drop of water ends in a different place, as they are part of formalized Dynamic systems
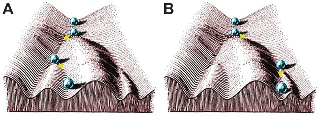

Source
Similarly, how can a zygote generate so many different cells if they have the same DNA?
Enzymatic systems of gene "on and off" switching
The existence of products (that later turned out to be proteins) in bacteria that regulate specific genes, was demonstrated by François Jacob and Jacques Monod in 1960 ⁶
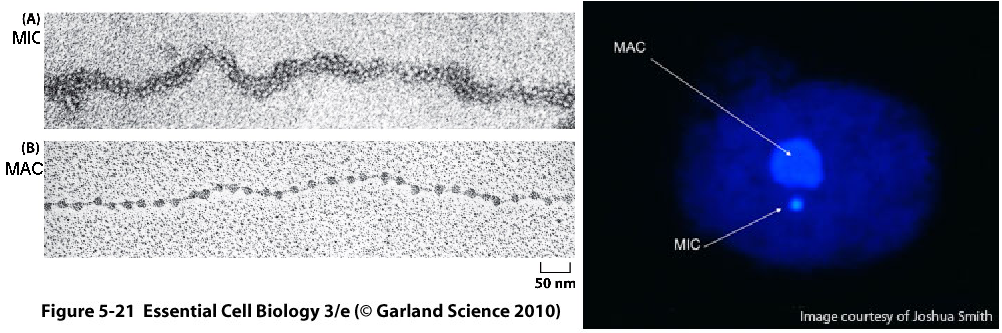
In 1964 Vincent Allfrey proposed that there would be chemical marks added to the tail domains that would instruct the cell's DNA to stay on or off.⁷ This required the need for opposing enzyme systems to add and take away the signal when needed. Later confirmed with the experiments in Tetrahymena thermophila a ciliated protozoan that posses nuclear dimorphism (One dense and one scattered nuclei). The molecular micronucleus (MIC) where DNA is densely packed is functional for sexual activity only and the macronucleus (MAC) is the one that is active during vegetative life.
Eukariotyc DNA is not naked, is packaged by histone proteins. Like knots, some are easier to untangle ((B) Euchromatin) than others ((A) Heterochromatin) and that information are more readily available for use.
As part of what is called the Adjacent Possible, in 1996 two Ph.D. students at different universities (Jim Brownell⁸; Jack Taunton ⁹), 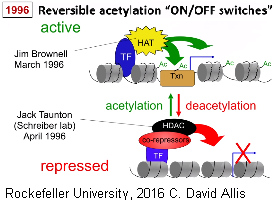 independently solved the two halves of this puzzle of enzymes HAT and HDAC. This is not the whole story as the enzyme readers, writers and erasers the acetyl groups is the on switch group. They are also transcription factors, which are the main protagonists in epigenetics.
independently solved the two halves of this puzzle of enzymes HAT and HDAC. This is not the whole story as the enzyme readers, writers and erasers the acetyl groups is the on switch group. They are also transcription factors, which are the main protagonists in epigenetics.
The silencing system foreseen in Alfrey studies appeared in the form of studies in Post effect variegation where methylation HMT ¹⁰ is the silencing signal (most of the time) and demethylation HDM ¹¹ the elimination of the silencing signal. As shown by Jenuwein and Shi respectively. 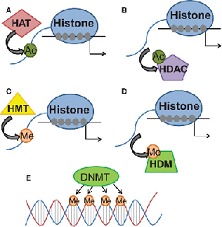
Source
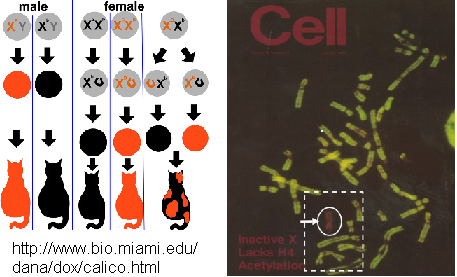
As show by chromosome X inactivation, hypoacetylation entails low transcription acivity.¹² as seen in the immunofluorescence picure where the green stain is lacking in the X chromosome.
The histones where the acetilation and metilation precesses are held are a rich field of study at the moment.
Out of the 5 histone proteins, H3 is the most post-translationally modified. Of particular relevance is the tail domain of the H3 histone complex, conserved form yeast to man, since they are a place for covalent modification and the 7 known variants seem to play a dynamic rol in the regulation of genes long term. ¹³
An interesting problem is that even when the active sites of histones are eliminated through mutation in yeasts it has not been found that they alter the DNA transcription and the resulting phenotypes, which only increases the doubts that they are relevant substrates even for the enzymes that modify them (histones).¹⁴
Targets for Medical Intervention
This is my favorite part of the subject. Maybe I'll make a specific post about it later.
One interesting consequence is that if a disease is secondary to epigenetic alterations, the damage is reversible. An example of this is Vorinostat a type of Histone Acetylase Inhibitor. This opens the door to treatments (not precisely cures) to altered cell lines. Which is just delightful. ❤
The blurred lines of controversy
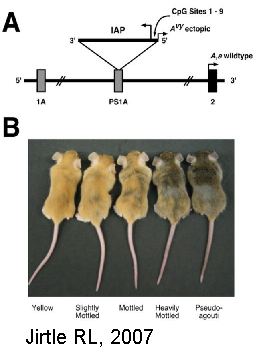
The agoutii mice experiment, where genetically identical mice are phenotypically different depending on gestational diet, If they are fed a methyl group rich diet get a clear fur, fat and bigger.¹⁵ This effect is lost by the third generation.¹⁶ This brings posibilities and hypothesis for modifications of sperm and egg cells during critical periods or the posibility that some of this changes can be transgenerationally permanent.
Transcription Factors
10% of all the proteins coded in the DNA (2600) have a function of reugulating the expression of genes, sometimes even RNA, by positive feedback mechanisms during critical periods. This means that unlike acethylation and methylation they can mantain their state after a unique non permanent contact with the activators. ¹⁷
An important example of this is the Yamanaka experiment on Induced pluripotent steem cells. Where a change in the cell identity is done by the induction of genes encoding transcription factors that act as DNA-binding regulatory proteins and transform adult cells into iPS and any histone modification seems to be secondary to this. ¹⁸
Maybe there's more than one way to skin a cat so to speak, but so far the evidence for transgenerational effects by modification of histones is lacking and seems redundant.
References
1 Allis CD, Jenuwein T.The molecular hallmarks of epigenetic control.Nat Rev Genet. 2016 Aug;17(8):487-500.
2 Pennisi E. A low gene number wins the GeneSweep pool. Science. 2003;300:1484
3 Oswald T. Avery, Colin M. MacLeod, Maclyn McCarty. STUDIES ON THE CHEMICAL NATURE OF THE SUBSTANCE INDUCING TRANSFORMATION OF PNEUMOCOCCAL TYPES. Journal of Experimental Medicine Feb 1944, 79 (2) 137-158
4 Heard, E., & Martienssen, R. A. (2014). Transgenerational Epigenetic Inheritance: myths and mechanisms. Cell, 157(1), 95–109.
5 Waddington, C. H. 1942. Canalization of development and the inheritance of acquired characters. Nature 150(3811):563–565.
6 JACOB F, PERRIN D, SANCHEZ C, MONOD J.. Operon: a group of genes with the expression coordinated by an operator.C R Hebd Seances Acad Sci. 1960 Feb 29;250:1727-9.
7 Allfrey, V. G., Faulkner, R., & Mirsky, A. E. (1964). ACETYLATION AND METHYLATION OF HISTONES AND THEIR POSSIBLE ROLE IN THE REGULATION OF RNA SYNTHESIS. Proceedings of the National Academy of Sciences of the United States of America, 51(5), 786–794.
8 James E Brownell, Jianxin Zhou1, Tamara Ranalli, Ryuji Kobayashi, Diane G Edmondson, Sharon Y Roth, C.David Allis. Tetrahymena Histone Acetyltransferase A: A Homolog to Yeast Gcn5p Linking Histone Acetylation to Gene Activation. Cell Volume 84, Issue 6, 22 March 1996, Pages 843–851
9 Jack Taunton, Christian A. Hassig, and Stuart L. Schreiber. A Mammalian Histone Deacetylase Related to the Yeast Transcriptional Regulator Rpd3p. ScienceNew Series, Vol. 272, No. 5260 (Apr. 19, 1996), pp. 408-411
10 Aagaard L, Laible G, Selenko P, et al. Functional mammalian homologs of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. The EMBO Journal. 1999;18(7):1923-1938.
11 Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (Dec 2004). "Histone demethylation mediated by the nuclear amine oxidase homolog LSD1". Cell 119 (7): 941–53.
12 Jeppesen P1, Turner BM. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression.Cell. 1993 Jul 30;74(2):281-9.
13 Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 2006 Jan 6;281(1):559-68.
14 Mark Ptashne1. Epigenetics: Core misconcept. PNAS April 30, 2013 vol. 110 no. 18 7101-7103
15 Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007 Apr;8(4):253-62.
16 Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals.Nat Rev Genet. 2012 Jan 31; 13(3):153-62.
17 Pomerening JR. Positive-feedback loops in cell cycle progression. FEBS Lett. 2009 Nov 3;583(21):3388-96.
18 Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006 Aug 25;126(4):663-76.

well my glip glop besides the massive owning of some dude with shaky arguments this whole cryptocurrency shitposting experiment has significantly improved your English grammar - which is significantly better than that of our spirochetes infested mutual friend - fact that I can tell from reading some of your first and last posts but is still not at a native level which means mine is not either... vis a vis - like that well dressed dude in the matrix said - mine must be currently in between yours and his... still distant from a native level... ergo the one thing I thought I was good at... I still suck balls at... I need another drink...
Check out my English skills.
I'm like some sort of grammar wizard.

very interesting :)