The Epigenetic effects of arsenic exposure.
The epigenetic effects of arsenic exposure
A short Life science review by @Clausewitz
If you enjoy biochemistry, genetics or science in general, I got something very interesting for you today, since I have put together a little life science review.
Here I will investigate the possibility of a unknown mechanism preformed by DNA methyl transferase 1, an enzyme responsible for maintenance methylation of DNA, leading to gene regulations from arsenic exposure.
I also intend to link the progression to nutritional factors as well as cancer progression.
Numbers in brackets e.g (5) means this is linked to reference nr 5 in the reference list.
.jpg)
Abstract
Chronic exposure to arsenic affects around 200 million humans worldwide and leads to many different health complications, amongst them cancer in different organs like liver, skin and kidneys.
Arsenic is a well known and well studied carcinogen, and arsenic oxides can even be used for cancer treatment.
There are many different cellular mechanisms that could be partially responsible for the progression of cancer making it hard to pin down exactly.
The recent concentration of studies on epigenetics and arsenic makes it a natural focus for this review and proves that scientists working on arsenic poisoning suspect that the mechanisms responsible for carncinogenesis will be found in this area.
The clear connections between, SAM depletion, DNMT1, and methylation patterns gives credit to these suspicions. Add the recently found inhibitory effects of iAs on CTCF, resulting in lost chromatin structure as well altered methylation status on histones, and the methylation patterns made by DNMT1 are certain to be affected.
Introduction
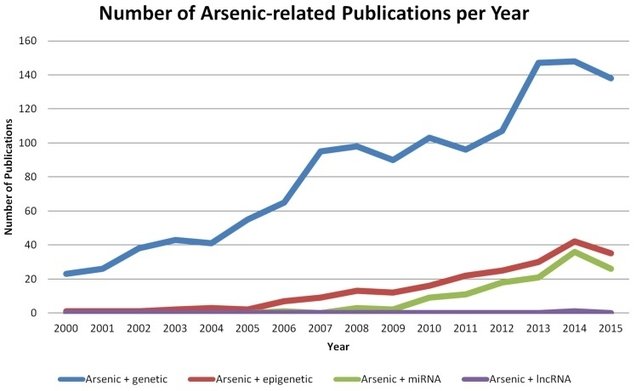
The number of studies on arsenic poisoning has increased fast and reached well over 100 publications in 2015 on the subject of arsenic and epigenetic regulations alone (Figure 1).
Earlier studies shows that arsenic is a weak factor when it comes to inducing mutagenesis, suggesting that mutations is not the primary explanation for cancer development after low dose, chronic arsenic exposure(1). Mutations, primarily deletions is first seen at direct toxic doses.
.jpg)
Figure 1: Number of studies on Arsenic and epigenetics during the period 2000-2015(20).
Arsenic depletes the methylation cofactor SAM
There is a growing body of evidence supporting that inorganic arsenic (iAS) interferes with important processes in cells such as methylation patterns in promoter regions and histones but also oxidative stress leading to wide cell damage, which historically is a more studied aspect.
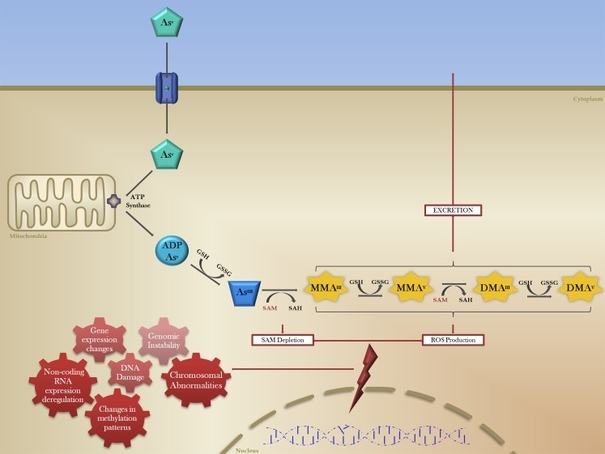
Chronic arsenic exposure is known to induce a state of global hypomethylation. When arsenic enters the human body, it is oxidised, bio-methylated and turned into mono-methylated As (MMA) & di-methylated As (DMA) (Figure 2). This process requires S-Adenosyl methionine (SAM) as a cofactor, just as DNMT1 and DNMT3 does, enzymes responsible for methylating DNA and carrying the correct methylation patterns to the next generation DNA after replication.
.jpg)
Figure 2: The bio methylation of arsenic. Inorganic arsenic is oxidized and later turned into Mono-methylated arsenic and di-methylated arsenic. The enzyme AS3MT catalyzes this reaction and uses SAM as a co-factor(19).
This process places a high demand on methyl levels in the affected cells leading to hypomethylation when the accessible amount of SAM drops. This is nothing surprising given the insight into the high SAM demand required in the bio-methylation process of As. SAM is also degraded to SAH which can have inhibitory properties for enzymes(20).
When the body's demand for SAM increases so does its production of SAM, primarily in liver cells. This increases the local concentration of SAM in liver and blood. Studies on blood samples from arsenic exposed patients seem to confirm that this is important. Paradoxically in these patients hypermethylation in many promoter regions has been repeatedly found, especially at CpG sites. Downregulation of cancer suppressor genes seem to be a common result in studies focused on these methylation patterns.
These results are more complex to explain mechanistically but it is known that CpG methylation, the methylation of cytosine-phosphate-guanine sequences, is preferred by DNMT1.DNMT1 or DNA-methyltransferase-1 is the enzyme responsible for preserving methylation patterns after DNA-replication pointing further to its possible involvement. THRF1 binds to chromatin, at CpG sites close to replication forks and helps in the recruiting of DNMT1 but also facilitates histone methylation, linking these pathways. Inhibition of CTCF, an important protein for chromatin structure and isolation, causes further destabilization of the chromatin so it must all be regarded.
Epigenetic regulations
Hypo & Hypermethylation patterns, CpG sites,UHRF1, CTCF, DNMT1, Histone H3 tail.
The secretion of arsenic is dependent on bio methylation by an enzyme called AS3MT, which uses SAM as a cofactor in this process, as seen in figure 2. The same cofactor is used by the enzyme group DNMTs, consisting of DNMT1,DNMT3a and DNMT3b(2).
DNMT´s are responsible for methylating DNA in vivo, both maintenance methylation and methylations during cell replication are carried out by DNMT´s.
When AS3MT and DNMT´s starts to compete for the SAM, it's available levels drop of, explaining the global lack of methylations seen, surprisingly enough alongside with gene specific promoter hypermethylation(3). Many studies have correlated global hypomethylation with cancer in e.g liver and prostate and seen cancer linked genes be upregulated, e.g HYAL1, short for the enzyme Hyaluronidase-1(4).
It seems that lower doses of iAs causes global hypomethylation and local, site specific hypermethylation. Studies on cell cultures only run for short time periods and use of different concentrations, often higher that what human cells would be exposed to. Niedzwiecki et al. did dose dependent study on humans PBMC DNA from patients with tested well water. The results confirm that more iAs can lead to more methylations locally within certain dose ranges(6). This creates a discrepancy in the studies since humans consume arsenic for years and the cells are exposed to much lower doses. Turning to studies on human cord blood clearly shows local hypermethylation patterns(6).
Pilsner.et al has reproduced these results, comparing cell culture to human leukocyte DNA in separate studies proving that hypermethylation are most common in human blood samples(18,7).
These studies also show a specificity of the hypomethylation status to CpG sites, Cytosine-phosphate-Guanine sequences in a longer DNA sequence, further pointing to the involvement of DNMT enzymes and UHRF1, a protein that bindes to methylated DNA in the S-phase of replication and helps with recruiting DNMT1.
Guangming et al showed decreased levels of P16 as a result of hypermethylation in the its promoter region(11)
Chanda et al showed similar results hypermethylation patterns were found at both the gene promoter of p16 & p53, both down regulated as a result(8,17).
P16 a protein is responsible for terminating the cell cycle between phases G1 and S, suppressing cancer growth.
The tumor suppressor protein P53 is knowns as a tumor antigen p53 and prevents genome mutations, so the lack of these to proteins would make it harder to prevent cancer progression in damaged cells
Further proving the dose and time dependency of creating hypermethylation instead of only hypomethylation is the studies by Cui et al who found dose dependent inhibition of DNMT´s and demethylation of CpG islands(9). He also,in a later study the same year, found hypermethylation corresponding to lowered concentration of p16 in lung cancer mice(10). The second study was conducted with very low doses over 18 months compared to the first study having higher doses but only spanning a 72 hour duration.The long duration required and advanced model systems needed, mice rather than cell cultures or best of all, human patients with known exposure, makes it hard to study these aspects directly. Known exposure on human patients hardly exists outside cancer trials which uses patients previously not exposed to arsenic or with unknown exposure, and the arsenic used , trioxide, are more toxic which also is seen in the study by Cui et al (9).
The longer studies with advanced model systems, mice or humans, seem to confirm hypermethylation resulting in down regulations of important cancer suppressive genes like p16 and p53. This concludes that arsenic can induce large epigenetic changes after long term chronic exposure in low doses > 100 ppm. It also concludes that hypermethylations can cause down regulations in gene expression of important cancer suppressive genes, linking it to cancer progression.
The most complex part is still to explain how DNMTs involvement and the CpG specificity seen creates hypermethylation patterns. Rea et al proved that iAs inhibits CTCF a protein involved in DNA binding and decreases its occupancy at the DNMT site by 40%. Another 20% reduction is gained from decreased expression which leads to lower levels DNMT (Figure 3)(12). CTCF is a zinc finger protein and it has been shown that monomethylated and dimethylated arsenite have higher affinity for zinc proteins, due to the high abundance cysteine residues(15,16) MMA and DMA is primarily seen in studies using low doses over long
time frames or in human patients and samples from human patients with low chronic exposure to arsenic. MMA & DMA production is known to take place during chronic low dose exposure which further links this state to deviations in epigenetic regulations.
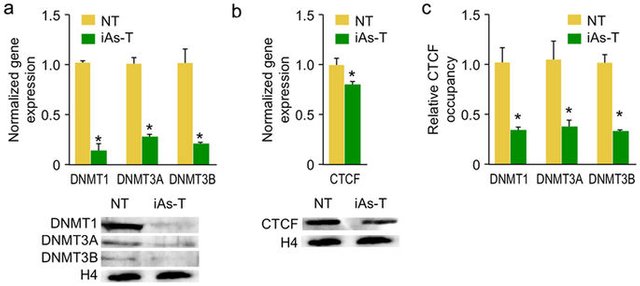
Figure 3: Expression levels of DNMT1,3a and 3b, clearly lowered during iAs exposure(9)
Arsenic is known to change methylation patterns at a histone level as well as at a DNA level, which was summarized by Eckstein et al.(13) This changes the chromatin structure and the accessibility to different genes.
DNMT1 known to bind to histones H3 tails, making the precision in methylation even more affected when histone methylation patterns change alongside the DNA´s.
This leaves lower levels of SAM, more SAH and fewer active enzymes, but in the liver, SAM production is up regulated leaving few active enzymes with access to more SAM. SAH is known to inhibit DNMT, a direct product in the bio-methylation of iAs, leaving even fewer active enzymes. In the liver SAH is turned to SAM again in a folate dependent process, giving the progression of diseases linked to chronic arsenic exposure a nutrition dependency as well. With sufficient folate and DNMT1 activity in the liver and blood the hypermethylation status of CpG regions might seem logical.
The active DNMT´s might have access to genes that were previously unreachable. However when the histone structure changes and decreased CTCF occupancy both changes the chromatin structure and regional insulation is altered. It is also likely that down regulations occur when histone and chromatin structure change as well as giving access to new genes.
This leaves it wide open for gene expressions to differ as the arsenic exposure continuous.
The H3 histone tail is important for DNMT1 to be able to bind to chromatin and methylate the intended DNA sequence.
DNA and histone methylation pathways were dynamically linked via protein UHRF1 an E3-ubiquitin-protein ligase, specialized in binding to CpG sites at replication forks, by Qin et al(14)
They also speculated in however DNMT1 can perform methylations other than maintenance.
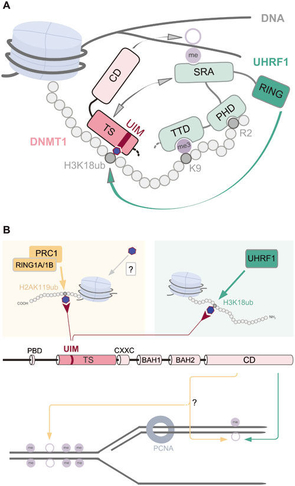
DNMT1 binding domains interacting with the histone H3K tail. UIM region binds directly to position H3k18ub seen in A.(14)
“The identification of the DNMT1 UIM now opens the possibility that ubiquitination of histone tail residues by ubiquitin E3 ligases might constitute alternative pathways for DNA methylation by DNMT1 CD beyond classic maintenance”. Quotation(14)"
Taken together, this could result in gene specific hypermethylation at CpG sites, but the general effect on histones and chromatin as well as DNA structure & repair might be just as important to regulating down genes that otherwise would suppress cancer. There could be proteins expressed that is not identified, involved in suppressing gene expression and the protein-protein interactions affected by arsenic needs to be mapped in more detail.
Concluding remarks
The time and model system needed to mimic the progression of iAs in the human body under low dose chronic exposure seems to be lacking in many ways and very few studies span over years with advanced models as mice or higher order animals. This puts a limiting factor on gaining insight into arsenic metabolism and the wide effects it has on epigenetic regulations.
In conclusion it seems very likely that DNMT1 is involved in the local hyper methylations in humans with enough folic acid, since it is required to replenish SAM levels. The lost structure of chromatin as well as lost insulation caused by CTFT inhibition affects gene expression. The same can be applied to histones when they lose structure, their tails become more available for binding to DNMT1. This point to that global hypo methylations can lead to local hyper methylations by DNMT1 via non maintenance methylation. Add the links to UHRF1 and UIM and I must conclude that it is more than likely that DNMT1 preforms methylations outside the priory known maintenance area.
Many epigenetic down and up regulations occur after arsenic exposure and the links to p53 & p16 links it to cancer progression.
The many possibilities for changed gene regulations could explain cancer progression by themselves, beyond the more precisely investigated links to p16 and p53 levels.
I made a Poster on Epigenetics and arsenic exposure  .
.
 .
.Reference list
1.Further evidence against a direct genotoxic mode of action for arsenic-induced cancer: Klein CB, et al. Toxicol Appl Pharmacol, 2007 . https://www.ncbi.nlm.nih.gov/pubmed/17316729/
2.S-adenosyl-methionine-dependent methyltransferases: highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications: Struck AW et al. 2012. https://www.ncbi.nlm.nih.gov/pubmed/23180741/
3.Differential DNA methylation profile of key genes in malignant prostate epithelial cells transformed by inorganic arsenic or cadmium: Pelch KE, et al. Toxicol Appl Pharmacol, 2015.https://www.ncbi.nlm.nih.gov/pubmed/25922126/
4.Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression: Zhao CQ, et al. Sci U S A. 1997 Sep 30 https://www.ncbi.nlm.nih.gov/pubmed/9380733/
5.Molecular events associated with arsenic-induced malignant transformation of human prostatic epithelial cells: aberrant genomic DNA methylation and K-ras oncogene activation: Benbrahim-Tallaa L et al. Toxicol Appl Pharmacol. 2005. https://www.ncbi.nlm.nih.gov/pubmed/16039940/
6.A Dose–Response Study of Arsenic Exposure and Global Methylation of Peripheral Blood Mononuclear Cell DNA in Bangladeshi Adults: Megan M. Niedzwiecki, et al. Health Perspect. 2013 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3855504/
7.Influence of prenatal lead exposure on genomic methylation of cord blood DNA: Pilsner JR et al. Environ Health Perspect. 2009 https://www.ncbi.nlm.nih.gov/pubmed/19750115
8.DNA hypermethylation of promoter of gene p53 and p16 in arsenic-exposed people with and without malignancy: Chanda S, et al. Toxicol Sci. 2006. https://www.ncbi.nlm.nih.gov/m/pubmed/16251483/
9.Arsenic trioxide inhibits DNA methyltransferase and restores methylation-silenced genes in human liver cancer cells: Cui X et al. Hum Pathol. 2006. https://www.ncbi.nlm.nih.gov/pubmed/16613325
10.Chronic oral exposure to inorganic arsenate interferes with methylation status of p16INK4a and RASSF1A and induces lung cancer in A/J mice: Cui X, et al. 2006 https://www.ncbi.nlm.nih.gov/pubmed/16543296
11.Arsenic exposure is associated with DNA hypermethylation of the tumor suppressor gene p16: Guangming Lu, Huiwen Xu, De Chang, Zhenglai Wu, et al. Med Toxicol. 2014 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4297462/
12.Genome-wide DNA methylation reprogramming in response to inorganic arsenic links inhibition of CTCF binding, DNMT expression and cellular transformation: Matthew Rea, et al. Scientific Reports https://www.nature.com/articles/srep41474
13.Reviews on Environmental Health: M.eckhart et al, 2016
14.DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination: Weihua Qin, et al. Cell Research (2015) https://www.nature.com/articles/cr201572/
15.Metallothionein-1 and -2 expression in cadmium- or arsenic-derived human malignant urothelial cells and tumor heterotransplants and as a prognostic indicator in human bladder cancer: Zhou XD, et al. Toxicol Sci. 2006 Jun.
https://www.ncbi.nlm.nih.gov/pubmed/16565513/
16.The metabolism of inorganic arsenic oxides, gallium arsenide, and arsine: a toxicochemical review: Carter DE1, Aposhian HV, Gandolfi AJ, Toxicol Appl Pharmacol. 2003 https://www.ncbi.nlm.nih.gov/pubmed/14678742/
17.Chronic arsenic toxicity: studies in West Bengal, India: Kaohsiung J, et al. Med Sci: Chronic arsenic toxicity: studies in West Bengal, India. 2011. https://www.ncbi.nlm.nih.gov/pubmed/21914522
18.Genomic methylation of peripheral blood leukocyte DNA: influences of arsenic and folate in Bangladeshi adults: Pilsner JR et al. Clin Nutr. 2007 https://www.ncbi.nlm.nih.gov/pubmed/17921400
19,20.Oncogenomic disruptions in arsenic-induced carcinogenesis: Adam P. Sage et. al. 2017.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5421966/figure/F3/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5421966/figure/F2/
Thanks for checking my work out, much appreciated, I hope that you gained some insight into the dangers of arsenic and the links to DNA-methyl-tranferases.

Nice payout ! your high on the list for curie and steemstem, congrats ,I repromoted again. I'm Jealous,
Thanks! They are simply the best @curie & @steemstem :) I am honored!
He's very brilliant at what he knows. and that's SCIENCE.
KEEP THE GOOD WORK ON, EXPECTING MORE SCIENCE WRITE UP FROM YOU
1 steem dollar put you 7 down on the 1st page of promoted. Lets see what happens.
Awesome, far to kind :) maby i should add some!
Another Epic Thesis, I feel like I just binge watched a month of jeopardy. :-o
I was wondering if you heard about MMS (sodium chlorate and citric acid) there are a lot of testimonials out about it.. They're saying its cures cancer and grows your hair back!
You're very right
I've been looking it up.
It's very promising,
thank you brother..
Thanks @Brotherdave haha yeah this might be a bit juicy, much biochemical terms, since i focused on the genetic & biochemical mechanisms this time :)
I'm checking out- you tube -steem upvoting bot tracker shows were to buy an upvote- we've been missing out. Every one of your juicy articles can be reposted on a bot with much better returns. also best post time is 10 am unless night owls are more likely to more responsive to your type of material ,which could be true. Anyway I'm giving the bots a go. Also did you look up Water Memory? very interesting.
Yeah it´s fascinating i am reading some controversial articles atm regarding water memory or related effects, might write about it in the near future :)
@Hardcorejokes Yeah i´ve heard it mentioned but seems worth looking closer at!
Appreciator has a lot of refund requests on his blogs. It's probably nothing , but was looking into it wasn't sure about participating
Seems to be working atleast
epigenetics is a hot topic. Great article. :) resteeming! :)
Yeah it is interesting indeed! Thanks for checking it out and thanks for the support :)
Real good article i can see you put lots of work. I know more about arsenic then before. Thanks for sharing
Thanks for reading, it sure is not safe to drink or eat :)
Really enjoyable read, keep posting good content. Epigenetics always excites me!
Glad you liked it and thanks for checking it out!
steemit 4 life!
Yeah it is the platform of the future :)
Great article!! Didn't know that arsenic had such a deep impact on the body..I'm trying to stop smoking at the moment, this another good motivator :) (tobacco is full of arsenic...and other crap)
Been wanting to comment on your other posts but I've been plagued by low bandwidth..but just love the way you finished off your post on telomeres. So obvious, yet its oblivious to most :)
'If you are trying to live longer, stop eating meat and pesticides'
Keep up the awesome work!!
Yeah tobacco got some sneaky additives, so keep going!
Yeah many humans eat crap and toxins along with alcohol and smoking then try to live longer with meds that enhance biological processes :) kinda contra productive !
Thanks a lot for checking my work out and the kinds words, I am honored :))
Peace
Haha I think psychologically we need the contradiction - a sense of freedom that we can do whatever, but at the same time we want to stick to higher principles. And all in the belief that these concepts are empirical facts

That is true, an never ending conflict of interest :)
Releasing your post on the weekend is kinda like swimming in a shallow creek. Even if we paid to promote it there's little flow.
haha :) why is weekend bad : / ?
There is so much to think about!