Single Nucleotide Polymorphisms: Single Letter Changes in DNA Provide Road Signs to Map Human Traits
Single nucleotide variations that are routinely found in the human genome DNA sequence can be used to identify regions important to understanding diseases.
The human genome sequencing project has culminated with the determination of the linear sequence of the approximately 3 billion bases of human DNA. Within this DNA sequence are the roughly 30,000 or so genes that encode the products that make humans human. Obviously there are gene differences between individuals but, perhaps surprisingly, all humans have DNA sequences that are 99% identical.
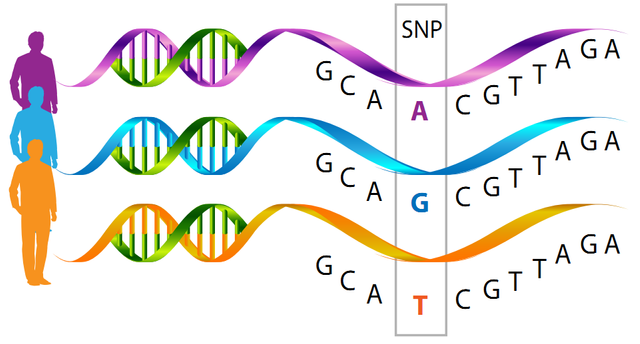
What Are SNPs
When the DNA from a particular region of a chromosome, say from chromosome 21, is sequenced from multiple individuals and the sequences compared, changes in a single DNA base between different individuals are found. These differences are referred to as polymorphisms (from the Greek meaning “many forms”). Since the change was found at a single location it is called a “single nucleotide polymorphism”, which is called an “SNP” (pronounced as ‘snip’).
What Do SNPs Tell Us
Single nucleotide changes in the human population DNA sequence are found in roughly 1% of all the bases of DNA. Only roughly 5% of the entire DNA in our genomes is responsible for directing the synthesis of proteins and other gene products. When a SNP is found in the genetic coding information for a protein it can indicate a change in the ultimate sequence of the protein, which may suggest a subtle, or sometimes not so subtle, change in the protein’s behavior.
Using SNPs as Mapping Tools
The modern era of DNA analysis has made the ability to rapidly characterize DNA sequences a far more tractable problem than ever before. Once a SNP has been found in a particular gene sequence, methods for detecting it in rapid fashion can be utilized. If a scientist hypothesizes that the particular gene may be linked to a particular disease, they can use the SNP as a marker. By comparing the DNA from a number of individuals with the disorder in question to a group of individuals without the disorder, they can see if there is a higher frequency of finding the SNP in people with the disorder as compared to those without it. Using statistical tools, it can be determined if the SNP is a significant predictor of the presence of the disorder.
Measuring More Than One SNP at a Time
Trying to associate a particular SNP with a disease can be a difficult problem. At present more than 1.4 million SNPs have been identified in the human genome. New technologies utilizing methods such as microarrays, polymerase chain reaction and mass spectrometry have made it possible to rapidly identify multiple SNPs in a single sample; yet new methods for large scale, robust SNP identification are still needed.
What Can SNPs Tell Us
Since most diseases are or can be influenced at some level by our genes, identifying multiple SNPs that associate with a higher risk of a disease could be useful, for instance, in beginning preventative measures or behaviors earlier. Also, SNPs can be used to identify particular people in a population that might respond better to a particular drug treatment for a disorder. The ability to tailor specific treatments to specific individuals while minimizing the risk of using a less effective treatment option makes SNP mapping an important tool for the future.
Source: