Journal of Unpublished Results – A case for the validity of scientific research (Part II)
Hello my scientific affiliates and science-skeptics of Steemit,
as I promised in the first part of my article about the reproducibility of valid science, in this second part, I am going to show you the results of my own experiments related to C/EBP-induced transdifferentiation of B cells, which was first described by Thomas Graf and colleagues in 2004. You should browse through the first part if you haven't read it or are not well familiar with molecular cell biology
The experiments
In my experimental model, I am using mouse B-lineage lymphoma cells (cancer cells of the immune/lymphatic system) that express an oncogene (i.e. a cancer-causing gene, let's call it X) in a B-cell specific manner. The expression of oncogene X is, at least partly, driven by another protein that we have already encountered in the previous post– Pax5.
I had to test whether the C/EBP proteins would also induce transdifferentiation of my particular B lymphoma cells, let's call them oncoX-cells. If I knew that this would work in my experimental system, I could perform the actual experiments that I was interested in. As mentioned in Part I, here I will only cover my test experiments.
I am going to refer, where appropriate, to the figures of the freely accessible, original publications and present my own results that are directly related to them. Please consider that I didn't just copy the published experiments. Instead, I was following my own logic, but in the end it turned out to be pretty analogous to what was shown by others.

What I did in the first step, was taking my oncoX-cells and genetically modify them to express C/EBPα or C/EBPβ. I will only show results obtained with C/EBPβ, but very similar results were obtained with C/EBPα.
1. Molecular markers
After I activated C/EBPβ for three days in the oncoX-cells, the first task was to analyze the cells for a few important cell surface proteins by a method called flow cytometry, in conjunction with fluorescently-labelled antibodies. These cell surface proteins are markers for the particular identity of the immune cells in question. This analysis would give me clues as to whether there are any changes in cell type caused by C/EBP activation.
Here I am showing x-y plots for four surface proteins: CD19 (a B cell marker), CD11b (a general myeloid marker), F4/80 (Monocyte/macrophage marker) and M-CSF-receptor (a receptor for the cytokine M-CSF that signals monocytic cells to undergo differentiation).
These results are related to figures 1 and 4 of publication (1). Note that Mac-1 designates the same protein as CD11b.
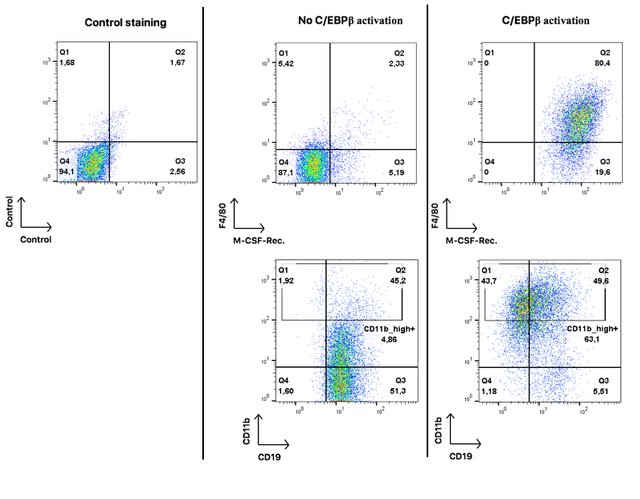
Each dot on these plots represents a single cell. On the very left hand panel you see a control plot to estimate the percentage of false positive results. The quadrant gate separates double-negative from both single-positive and double-positive events.
In the middle panel you see oncoX-cells without activated C/EBPβ. They are mostly negative for F4/80 and M-CSF-Rec, positive for CD19, and only about 5% highly positive for CD11b. I should add that I used an extremely sensitive antibody for CD11b and it is not unusual for B cells to express some mid-range level of that protein (pay attention to the logarithmic scale of the axes).
On the right hand panel, you see oncoX-cells that have expressed C/EBPβ for three days. All of them acquire the expression of the M-CSF-Rec., a large proportion starts to express F4/80 and high levels of CD11b while about half of the cells already loses the expression of the B cell marker CD19.
These results imply that oncoX-cells that were forced to express C/EBPβ acquire surface proteins that are indicative of myeloid differentiation and at the same time loss of B cell identity. Some of the cells (the CD19 and CD11b_high double-positive) have a mixed cellular phenotype at this point.
Now, you can say that these are just some colorful plots and you are not convinced. You would be right to do so. I am going to show you very illustrative material.
2. Induction of differentiation - View under the microscope
One of the features of immature myeloid cells is their ability to respond to myeloid-specific signals, such as certain cytokines. Two of the most important cytokines for myeloid development are M-CSF (macrophage colony-stimulating factor) and GM-CSF (granulocyte-mactophage colony stimulating factor). I need to add that C/EBPβ activated oncoX-cells not only start to express the M-CSF receptor as shown in Figure 1, but also start to express the GM-CSF receptor (flow cytometry data not shown).
So I wondered what will happen if I supply my C/EBPβ activated oncoX-cells with these cytokines. The results are somewhat related to Figure 3 in (2), although these papers have added M-CSF to their cultures from the beginning and thus have not explicitly tested the propensity of their transdifferentiated cells to react to B cell foreign cytokines.
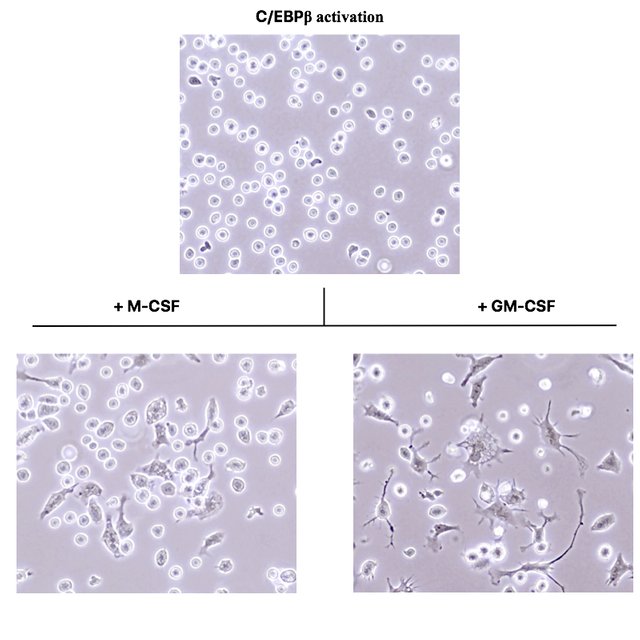
These photographs were acquired under a phase contrast microscope with a 400x magnification.
As you see, the C/EBPβ activated cells in the top picture are still round and do not morphologically differ from normal oncoX-cells. However, after just three days of culture in presence of the cytokines M-CSF and GM-CSF, many of the cells drastically change their morphology and adhere to the culture dish bottom.
While cells exposed to M-CSF assume a morphology very reminiscent of macrophages (compare to Fig. 3 of (2)), cells exposed to GM-CSF assume a morphology identical to dendritic cells. (I verified this by taking normal mouse bone marrow and culturing it for a week in presence of these cytokines.)
If you leave the C/EBPβ activated oncoX-cells for a week in presence of these cytokines, you even get a complete conversion, i.e. there are hardly any round and floating cells left. This is something that will never happen with genetically untouched oncoX-cells, no matter how many myeloid cytokines you put on them.
3. Testing transdifferentiated and further differentiated cells for phagocytic function
In addition, I needed to test if the morphological changes elicited by the cytokine cues would correlate with phagocytic function - because that's what macrophages and dendritic cells do: they take up specific and unspecific molecules, particles or even microbacterial organisms from their environment. They literally eat them!
I supplied C/EBPβ activated oncoX-cells with fluorescently-labeled Escherichia coli bacteria. These particles would lighten up only if they are internalized by the cells (due to the acidic environment in phagolysosomes). This is how it looked under a fluorescence microscope, in accordance with Figure 3A from (3).
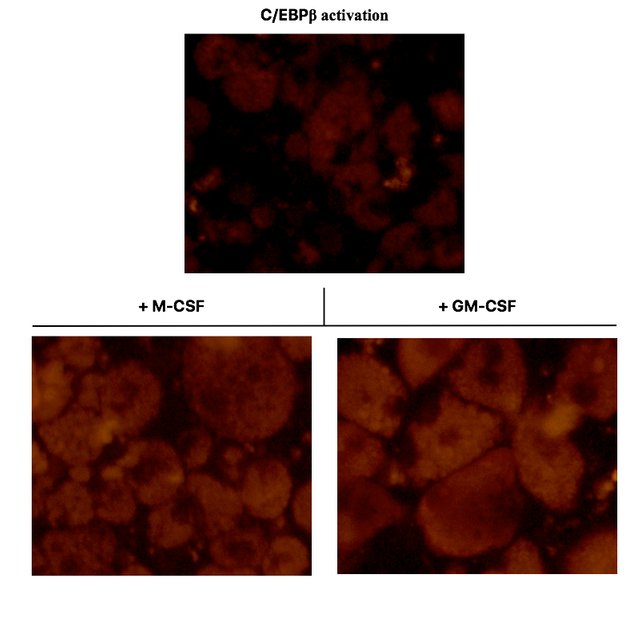
As you can see in the top panel, C/EBPβ activated oncoX-cells, which also to a small fraction spontaneously differentiate to macrophage-like cells, only yield a relatively weak fluorescent signal when fed with E. coli particles. The majority of C/EBPβ activated oncoX-cells hardly phagocytose at all and are not visible in this fluorescence setting./p>
However, culturing the cells with either M-CSF or GM-CSF impressively boosts their phagocytosis capacity and quantitatively leads to most of the cells acquiring phagocytic function, as can be seen in the bottom panels. Pay attention to the visible vacuoles representing the plentiful of phagolysosome organelles within the cells. The nuclei of the cells remain dark.
4. Examining gene expression changes
Lastly, I wanted to know what happened to the gene expression of B cell specific transcription factor Pax5 and the oncogene X that is also expressed in a B cell specific manner. Please refer to Figure 6D of (2). This is the result I got when performing mRNA quantification analysis:
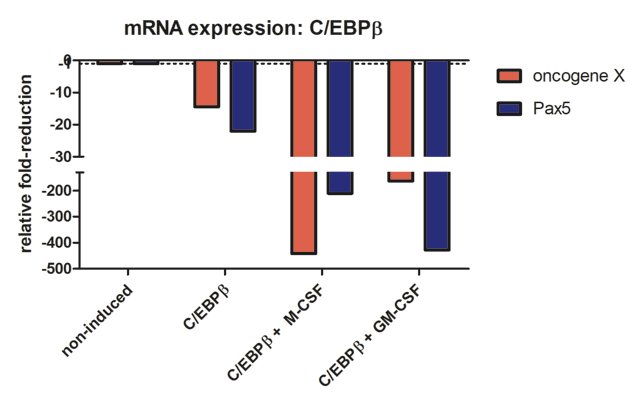
Compared to oncoX-cells without C/EBPβ expression (non-induced), C/EBPβ activated cells show marked reduction of both oncogene X and Pax5, while additional culture with the cytokines for seven days leads to further drastic reduction in the expression of these genes, correlating with the dramatic change in cellular phenotype. It should also be added that these cells stop dividing, in agreement with Figure 3 from (3).
In conclusion, enforced expression of C/EBPβ or C/EBPα in mouse B lymphoma cells leads to an impressive change of their cellular identity - this process is called transdifferentiation. Furthermore, the transdifferentiated cells acquire the ability to respond to myeloid cytokines with subsequent differentiation. Finally, the reprogramming of oncoX-cells ablates their primary oncogenic/proliferative driver, which basically means that they lose their cancerous features.
Shortly, with one relatively simple genetic trick I managed to completely disarm and change the behaviour and looks of some pretty evil cells that would usually kill a mouse within 2-3 weeks!
All of this is in accordance with publications (1), (2) and (3).
I hope, I could demonstrate to you that truly valid scientific research is easily reproducible by other, unrelated scientists and even under different experimental conditions.
This is the amazing thing about science. Once a fundamental principle is discovered, it can be tested in other settings, verified and expanded upon by scientists all over the world.
Of course, accomplishing this requires some intellectual, material and logistical resources, not easily available to just anyone.
EDIT You can use the tag "junpubres" (Journal of unpublished results if you want to contribute :-)
Yours, @replichara
References:
(3) Rapino, F., Robles, E.F., Richter-Larrea, J.A., Kallin, E.M., Martinez-Climent, J.A., Graf, T. (2013) C/EBPα induces highly efficient macrophage transdifferentiation of B lymphoma and leukemia cell lines and impairs their tumorigenicity. Cell Reports. April 25, 3(4):1153-63.
Image Sources:
-1- Pixabay
-2- to -5- own data graphs
Beautiful flow cytometry! It always makes me happy when I can follow along with others' work. Looking forward to more in this series. After the events of this workweek, I am planning on starting a series of my own, on how a total newbie can learn to claw and bite their way through genome mining.
Thank you for the approval!
I haven't dived into the bioinformatic side of the next generation sequencing technologies so far, so I am looking forward to learn something about it.
It is...challenging to say the least. Probably the most challenging part is convincing IT that yes, I need admin privileges, and yes I really do need Linux installed on my machine!
Haha... Yes, that's one the annoying things, to not be able to simply install software that you need to be able to work.
This is probably the realest-looking science I ever saw on steemit!
We need to show steemit that science can also be real, just like travel photos, haha!
Hi there,
another researcher is tolking here, I'm from Italy but now I'm working in Spain on Bladder cancer samples.
I'm performing GES ( gene expression sequencing) serching for a gene signature that will be usefull in diagnosis and prognosis of some bladder cancer patients.
I would like to thank you for your article, but expecially for what you did. The thing that the scientific results should be reproducible often is taken for granted, but find a researcher who spend his time to really reproduce it, is uncommon.
My compliment to your work and to you article.
Thanks for reading and appreciating the article! Yes, it is great when you can reproduce certain findings. It gets more difficult if you cannot, because you have to exclude that you did not do any conceptual, methodological or logical mistakes first, before really questioning previous research.
So you are working with one of the big data tools :-) Are there already bladder cancer subtypes defined based on gene expression signatures or are you at the frontiers of this approach?
there are alredy subtypes, there are only four important studies and in 2 of these they "mixed up" invasive and non invasive tumors to search the subtypes. My research will consider only the invasive tumors, and we are going to search for genes with a relative usefullness also for an immune therapy
I remember that our body in the dissection course had its bladder completely filled out by a giant tumor, so the doctors had to put a catheter into the ureter for urine to flow out. That was awful...
I wish you success in finding something distinguishing!
I reallly love the " on the field" part too!!
Great scientific approach. And I like the idea of a journal of unpublished results. It was very hard for me to get my doctoral theses accepted because I was ahead of times and it took 10 years to explain the idea to other doctors...
Hard work will only get you so far. Perseverance and belief in your own abilites are what will determine your success, I guess!
Educative. Will follow you to always learn from your subsequent post.
I will try to not disappoint you :-)
Nice research! Well done
Thanks :-)
Hola te invito a pasar por mi blog y leer mi ultimo post el embarazo se contagia !! @amirjv1 saludos
Sorry, I don't understand what you are saying.
Please vote for my articles I need some cash for my tuition fee pls
No.