Hope For The Fight Against MRSA: We May Be Able To Get Around Antibiotic Resistance By Altering The Bacterial Surface
I have written a few articles about antibiotic resistant bacteria before (you can read them here and here), so if you have been following my blog you are likely up to date on this growing problem.
In these previous articles I discussed with you all the need to develop new antibiotics, and my concerns over large pharmaceutical companies reluctance to prioritize this sort of research because antibiotics just don't bring in the kind of money that other medications do. This post will break down a new research article published in Nature Scientific Reports titled "Surface mediated cooperative interactions of drugs enhance mechanical forces for antibiotic action."
Lets Begin With The Conclusions Of The Article
- The experiments here show how important surface effects are in important to understanding how an antibiotic functions (in general).
- Significant steps were made here in understanding the mechanisms through which the cellular membrane surface layer regulates the interaction rates of antibiotics.
These conclusions may sound a bit science jargonish right now, but bear with me a moment and read onward. I will explain some of what the authors observed and help make the concepts in those conclusions make a bit more sense.
Lets Talk About The Bacterial Cell Wall
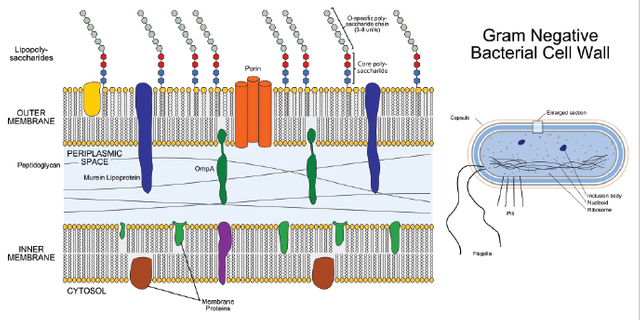
In the above image, we are looking at the cell wall for a gram-negative bacteria (there are two classes gram-negative, and gram-positive... I will go into this a bit more shortly). Now when you look at this you can see that the cell membrane is comprised of layers, the outermost layer is composed of "lipopolysaccharides" which are just sugar molecules bound to the phosphate head group of the phospholipid by layer of the "outer membrane."
Phospholipids are molecules with water loving head groups and water hating tails, the water hating tails contact each other in the by layer allowing the water loving head groups to contact water on either side (necessary for separating the water inside the cell, from the water outside the cell in its environment.
Also in this outer membrane we have proteins which span the membrane and allow for molecules to pass from the outside of the cell in, or inside of the cell out (these can be ion channels, or as is labeled in the figure porins
As we keep moving inward we reach the periplasmic space, or periplasm. In the periplasmic space of a gram-negative bacteria is a material called peptidoglycan, which is a mesh composed of amino acids linked together (this is called a peptide) which is covered with a variety of sugars. The periplasmic space also contains a jelly like material as well as a variety of proteins responsible for binding metals, amino acids and other molecules necessary for the survival of the bacterium.
Finally we reach the inner membrane which is another phospholipid bilayer and the final layer before the inner portion of the bacterial cell (where its DNA, ribosomes, metabolic proteins and all other cellular components reside!).
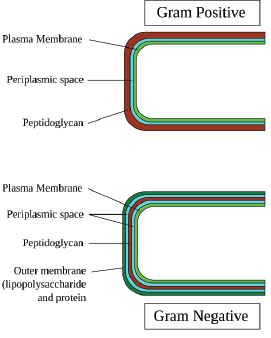
Now I mentioned to you that there are two classes of bacteria gram-negative and gram-positive. The cell wall we just discussed is for a gram-negative bacterium and it is the more complex of the two. The gram-positive setup is much more simple and is organizationally described in the figure to the right.
We can see that a gram-positive bacterium is basically missing the outer-membrane. It just starts with a pepdidoglycan layer, followed by the periplasmic jelly and finally an inner phospholipid bilayer membrane separating the outside of the cell from the inside.
Most Infectious Bacteria Are Gram-Positive
Okay now that we know a bit about what the bacterial cell wall is composed of, we should also be aware that most of the bacteria that we are concerned about fighting off happen to be gram-positive, including Staphylococcus aureus (of which you may be familiar with the antibiotic resistant variety known under the acronym MRSA).
So About The Article
As I said above and you are likely aware, antibiotic resistance is becoming a growing issue, and one in which you, your loved ones, and I may have to deal with at some point in our lives. What you may not know is that the way bacteria become resistant to many of the antibiotics we use is through changes that are made to the bacterial surface. These changes alter the ways that drugs interact with bacteria.
In this article the authors were looking for ways that the interactions between drugs which target bacteria that are resistant can be enhanced. To do this they looked at the forces (by force here I literally mean physical force) generated at the cellular surface by four different antibiotics:
vancomycin, ristomycin, chloroeremomycin and oritavancin
They found that there were different forces generated on the membranes of bacteria resistant to these antibiotics, that were not observed in organisms that are susceptible to them.
To measure these forces the researchers used a cantilever in which systems have been devised allowing researchers to measure forces as small as from a single hydrogen bond (Source. Using this method the researchers could measure added strain on a model for the cellular membrane (if there was any) when antibiotics were added.
Three Cell Membrane Models
The authors created monolayers composed of three different substances (polyethylene glycol (PEG) as a reference), a layer comprised of components found in normal bacterial cell membranes called vancomycin-succeptible receptor (VSR) (Source for VSR) which can interact with the antibiotic vancomycin, and a "reprogrammed" version of VSR where that interaction is weakened called vancomycin resistant receptor (VRR) (Source for VRR.
Looking For Effects on the Membrane Models
The first thing the authors did is looked at how much of the antibiotics were required to interact with the membrane models.

(a) What we are looking at here is the response of the injection of vancomycin against VSR (The succeptible membrane model). PEG was their control and you can see that nothing happens, however when using VSR in two different salt concentrations, and illustrates that a good response can be measured regardless of ionic strength.
(b) Here they were testing the thickness of the membrane and found that thicker membranes result in slower binding of the antibiotic.
(c) Here the researchers are looking at the binding of vancomycin to VRR (the resistant cell wall), which is small (it's the green line). They also looked at this when they have free VSR in the solution which not bound to their cantilever (here thats VRR) has on the mechanical response, what we see is that even low concentrations of the VSR in solution are able to bind up all the antibiotic and there is no longer a good response for vancomycin binding to VRR.
(d) This is the same experiment I described in C rather than vancomycin being added the researchers were adding a different antibiotic oritavancin (ori) , here they see a big response when the antibiotic binds (green), and a reduction in the size of the response when the free VSR is in the solution (purple and red). The reason the binding causes a big change here but not in C is because VRR is resistant to vancomycin binding, but not to oritavancin binding.

Now the researchers began to expand the scope of their experiments looking at all four mentioned antibiotics (vancomycin (Van), ristomycin (Rist), chloroeremomycin (CE) and oritavancin (Ori))
(a) here they were looking at CE on the VRR membrane, and showed that there is a drastic difference in response for this antibiotic just as they saw for Van previously. However they show a strong response for the VSR membrane.
(b) Showing that ALL antibiotics cause a strong response in VSR membrane
(c) shows that only Ori is able to create a strong response in the VRR membrane
(d) Shows the minimal concentrations of the antibiotics necessary to kill an antibiotic succeptible bacterial strain (filled shapes) versus resistant cells (open shapes). On the x-axis lower concentrations are on the left and higher on the right. What we can note from here is that the antibiotics that cause a strong response in their membrane force detection assays all have low inhibitory concentrations to cellular growth, but those that don't cause a response in the VRR membrane (Van Black, Rist-blue and CE-purple ) also don't kill resistant cells until very high antibiotic concentrations.
Why Is All Of This Important
So I just threw a lot of information at you, and you may be asking your self why is any of this important. Well here is why. The researchers found that in addition to the standard mechanisms by which these antibiotics are thought to work (stoping the synthesis of the peptidoglycan layer of the cell wall), the binding of the molecules themselves to the membrane causes strain. This strain seems to correlate with the ability of the antibiotic to kill bacteria.
AKA there is a second mode of action, that we did not know about before.
This opens up a whole new avenue for researchers, knowing this we can begin looking for compounds which will increase the mechanical stress of the membranes and potentially restore the effectiveness of these antibiotics at killing cells that are otherwise resistant to them.
TL;DR
Researchers have observed that in addition to their normal method of action, some antibiotics seem to destabilize the bacterial cell wall.
This destabilization appears to correlate to the antibiotics ability to kill the cells.
This discovery indicates that other compounds which are not antibiotics but can put stress on the bacterial cell wall, may be able to increase the effectiveness of antibiotics at killing cells which we currently consider resistant (aka with helper compounds, we may be able to fight MRSA with the antibiotics we currently use, but that currently don't work).
It's fascinating that we are still finding out new things about the mechanism of action of something as simple as an antibiotic, many of which have been in regular use for decades.
Sources
- http://www.nature.com/articles/srep41206
- https://steemit.com/science/@justtryme90/nevada-woman-dies-of-superbug-resistant-to-all-antibiotics-available-in-the-united-states
- https://steemit.com/science/@justtryme90/the-future-treatment-against-antibiotic-resistant-superbugs-may-be-inside-of-you-right-now
- https://en.wikipedia.org/wiki/Porin_(protein)
- https://www.boundless.com/microbiology/textbooks/boundless-microbiology-textbook/cell-structure-of-bacteria-archaea-and-eukaryotes-4/cell-walls-of-prokaryotes-34/gram-negative-outer-membrane-260-4739/
- http://www.nature.com/nnano/journal/v6/n4/full/nnano.2011.44.html
- http://www.nature.com/nnano/journal/v9/n3/full/nnano.2014.33.html
- http://pubs.acs.org/doi/abs/10.1021/bi00107a007
All Non Cited Images Are From Pixabay.com and Available Under A Creative Commons License
Any Gifs Are From Giphy.com and Are Also Available for Use Under A Creative Commons Licence
** Nature Scientific Reports is Open Access, and Re-use of Information and Images Available Under A Creative Commons License**
If you like my work, please consider giving me a follow: @justtryme90. I am a PhD holding biochemist with a love for science. My future science blog posts will cover a range of topics in the biology/chemistry fields.
Thank you for your continued support of my work! I appreciate all those who follow me (and those who don't too).
Good article. I was researching and reading about Super bug infection caused by NDM-1 gene due to Overuse of Antibiotics a day ago and i got much info from your article.
Whoops, two windows open. :D
I am glad you were able to get something out of it! Thanks for reading, and most of all for taking the time to leave a comment! :)
I like it.
@fahmiauliasfr upvote follow my posts, I am new in steemit. Thanks for helping. Congratulations and success
This is good news. I think your previous posts have given me a better background to understand what it means.
Thanks, going to keep at it. This particular article was interesting because it shows a new aspect to how antibiotics effect bacteria. With a new wrinkle to their effects it opens up the door to other compounds which could function in a synergistic way.
For as much anxiety (scientific anxiety at this point rather than real anxiety) over antibiotic resistant bugs, the amount of interesting new avenues for tackling it that I read give me hope that the issue will be solved before it truly grows much larger.
Upvoted, Followed and Re-steemed! Great work.
Thank you very much. Glad you liked it, and I appreciate both your comment and re-steem. :)
Great article thank you for sharing it. I am most interested in how cannabinoids can be used either a single agent antimicrobial or even improve efficacy of failing antibiotics through its "entourage effect."
Prof. Appendino added that the most promising cannabinoids in their study – cannabidiol (CBD) and cannabigerol (CBG) – also happen to be non-psychoactive.
https://www.ncbi.nlm.nih.gov/pubmed/18681481
https://www.ncbi.nlm.nih.gov/pubmed/19344127
I hope that people are working on these avenues. There is a lot that it seems various canabanoids can do, just need the research effort put into figuring out which are going to really work well, and which won't.
There is a lot of promising work out there that could help us tackle the issue of antibiotic resistance in bacteria before the problem grows large enough to really have an impact on a lot of peoples lives.
thanks
Thank you for reading!
This post has been ranked within the top 25 most undervalued posts in the first half of Feb 06. We estimate that this post is undervalued by $9.95 as compared to a scenario in which every voter had an equal say.
See the full rankings and details in The Daily Tribune: Feb 06 - Part I. You can also read about some of our methodology, data analysis and technical details in our initial post.
If you are the author and would prefer not to receive these comments, simply reply "Stop" to this comment.