How I Analyze DNA with Biopython
With its highly readable syntax and uncluttered visual layout, Python is one of the most versatile and adopted programming language. Many programmers can ignore that Python is not as fast as lower level languages when they have the modularity and the ability to scale things faster. Plus, the community is nothing short of amazing.
Speaking of modularity, in Python there's a module for everything. Whether you want to do complex math, statistics, machine learning, text processing and text recognition, penetration testing and even bioinformatics, there's at least one module you can work with.
In fact, my interest in bioinformatics is what allowed me to go over the struggle of learning to program. I tried to teach myself coding several times and I failed every single time. It was earlier this year when I started to take off from the ground and it was because of my active interest in genomics and bioinformatics.
This is how I discovered Biopython, a Python module. With something specific to practice on and to play with everyday, I started having a better understanding of both worlds.
In this post I will show you some basic stuff you can do with the Biopython module; and if you're interested you can learn on your own from here on. In future posts I will discuss other interesting/helpful Python modules.
Playing with DNA - The Biopython Way

While there are several modules geared towards bioinformatics, the one that I learned and practice with is Biopython. It was my best choice because I found several books discussing it at length, one of these books being the free Biopython Cookbook that you can read here. It's like a hands-on approach, 400 pages long. I've inspired the following examples from the book.
Before getting into the nitty-gritty I'm going to make a couple of assumptions:
- you have some knowledge of biology, especially genomics
- you have some knowledge of computation and programming languages
I am not going to explain genomics and DNA sequences, what Python is or how to install it because the post would turn to be unbearably long. There are countless tutorials/lectures/videos freely available out there for that purpose. Here I'm trying to be very specific:
- how you can use Biopython in computational biology.
According to the Cookbook:
"The Biopython Project is an international association of developers of freely available Python tools for computational molecular biology."
To install this module, go to the download page here. Make sure to install dependencies before you install Biopython.
I use Biopython in Windows, but you can use it in Mac or Linux as well. In Windows, for easier installation, you could get the 'wheels' from here (dependencies and latest Biopython) and install them via pip.
Now I'll show you some stuff you can do with it.
1. Instead of making up a DNA sequence, I'll take a real genetic sequence from the National Center for Biotechnology Information (NCBI):

- I'll click on Fasta
- copy the sequence:
CTGATTGGCCACTCTTCACGGGGGTAATATTAAATGGTCTCCCGCTATACTATGGGCTTTGGGATTCATTTTCCTATTCACCG
TAGGGGGCTTAACAGGAATTGTATTAGCAAACTCCTCATTGGATATTGTCCTTCACGACACATACTACGTAGTAGCCCACTTC
CACTACGTCTA
- alternatively I could save it as Fasta or Genbank file and work with it directly. Biopython knows both extensions.
2. In Python
- importing Biopython Seq and Alphabet methods
- inputing our sequence:
>>> from Bio.Seq import Seq
>>> from Bio.Alphabet import IUPAC
>>> myseq = Seq('CTGATTGGCCACTCTTCACGGGGGTAATATTAAATGGTCTCCCGCTATACTATGGGCTTTGGGATTCATTTTCCTATTCA
CCGTAGGGGGCTTAACAGGAATTGTATTAGCAAACTCCTCATTGGATATTGTCCTTCACGACACATACTACGTAGTAGCCCACTTCCACTACGTCTA',
IUPAC.unambiguous_dna)
3. Let's find the complement sequence of myseq:
>>> myseq.complement()

You can see it shows truncated (not in its entirety). You could use the print function to see the full sequence:
>>> print(myseq.complement())
- similarly, you find the reverse complement with
myseq.reverse_complement()
4. Assuming that myseq is a coding sequence, let's transcribe it to messenger RNA.
According to the cookbook:
"The actual biological transcription process works from the template strand, doing a reverse complement (TCAG ! CUGA) to give the mRNA. However, in Biopython and bioinformatics in general, we typically work directly with the coding strand because this means we can get the mRNA sequence just by switching T ! U."
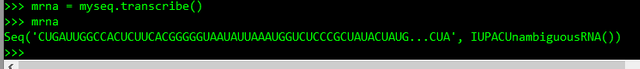
>>> mrna = myseq.transcribe()
>>> mrna
5. Let's translate the messenger RNA to protein:
>>> mrna.translate()

5. Parsing full records
- search the Entrez databases (like Pubmed, Genbank, Nucleotide, etc) with
Bio.Entrez.esearch():
>>> from Bio import Entrez
>>> Entrez.email = "[email protected]"
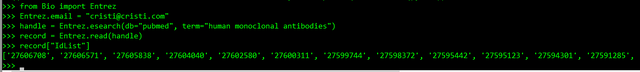
>>> handle = Entrez.esearch(db="pubmed", term="human monoclonal antibodies")
>>> record = Entrez.read(handle)
>>> record["IdList"]
And you get a list of Pubmed IDs of articles related to your search:
- you could retrieve the full entry for an id with
Efectch:
>>> handle = Entrez.efetch(db="nucleotide", id="186972442", rettype="gb", retmode="text")
>>> print(handle.read())
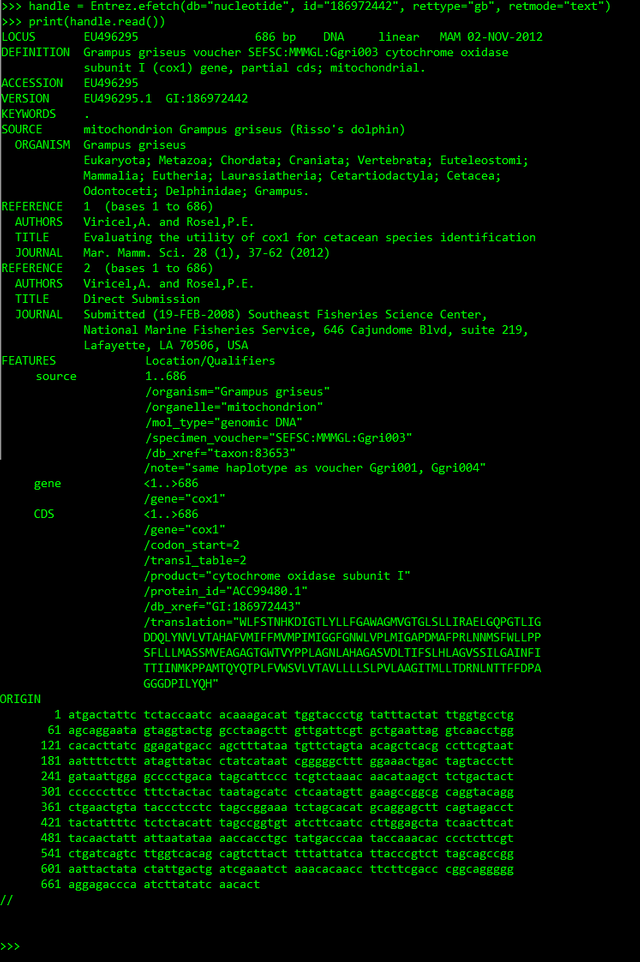
Here I retrieved the id 186972442 from the nucleotide database as Genbank file (rettype = 'gb' and retmode = 'text').
What else can you do with Biopython?
- find the lineage of an organism
- run BLAST (basic local alignment search tool) with
Bio.Blast - work with Swiss-Port and ExPaSy
- 3D protein rendering with the PDB module
- Population genetics
- Phylogenetics and Sequence motif analysis
- machine learning (!!!)
- and many other useful operations for bioinformaticians and computational biologists.
For this tutorial I used the Cookbook version from December 2015. The latest (which I linked above and here - the pdf) is from August 25, 2016. So, it's 'extremely' recent.
Ending Thoughts
If you want to follow the same learning curve as I did (genomics + programming), there are a few free resources that I'm going to provide to get you started. These are the ones that I used:
- Introduction to Biostatistics and Bioinformatics from NYU
Course Materials: here
My favorite lecture: IBB 4: Biopython
- Dr. Martin Jones' books:
Python for Biologists
Advanced Python for Biologists
- Via, Rother, and Tramontano's book:
Managing your Biological Data with Python
- there are a few other books that I wish to go through next (on my watchlist), but I'm only going to mention one:
Haddock and Dunn - Practical Computing for Biologists
Of course, you can watch many other free lectures on Youtube and go through course materials from top universities.
The only thing that matters, in my opinion, is to actually do the work. Start with 1 book or 1 course and get your hands dirty. Practice deliberately and do it consistently.
To stay in touch, follow @cristi
#bioinformatics #programming #genomics #research
Credits for Images: Dna Strands via Wikimedia Commons and Biopython Logo.
Cristi Vlad, Self-Experimenter and Author
You'll probably be interested in my (rather underappreciated) how to compare gps tracks post.
you know, I check the bioinformatics tags :) and I saw you posted it a few weeks ago.
Awesome post, thanks for sharing. I can't wait to see what the future of bioinformatics looks like!
Thank you for sharing this material, I like what you posted. Thank you so much
what do you like about it?
Interesting application of python, didn't know this existed. I don't know if it would save me any time over any of the available internet tools though. Will think about looking more into this. Thanks for posting about it @cristi!
it could save time if you script things, not doing them in the interpreter like I did here :) think about it.
This is soooooo good, loved this post, followed four times if i could lol
shall I consider this a troll post?
Four times maybe? :)
Fantastic post. I really must learn at least one programming language. Would Python be the best one to start with?
any kind of programming language is good to start with. for me it was python, as I said in the end, what's important is to code everyday, consistently!
Python is indeed very powerful. In particle physics, many largely used software have a pythonic interface and a C++ or Fortran core.
now that I have that particle physics books maybe I can find some applications to work with .
I can actually show you what I am developing with a colleague, if you are interested. That's an 'LHC analysis made easy' pythonic tool, that is actually used a lot by high-energy physicists (and we are missing developers). And let's stop the advertisement here ^^
dont get me excited! :D really, I'm interested.
I was actually thinking about writing a post about the idea behind this program. I will take your comment as a further motivation. When the post will be written, maybe could we then discuss about it on the chat.
What do you think?
Im up for that